|
To view this email as a web page, click here. |
 |
|
Welcome
We outline the procedure to create Quantitation Summaries for large data sets.
This month's highlighted publication offers a new approach to identify more host cell proteins in biopharmaceuticals using native digestions, Protein A and FAIMS.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to clear a strange error message in Database Manager.
If you missed our June newsletter, you might like to know that all the presentations from our user meeting at ASMS 2020 Reboot are available online as videos or PDFs.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
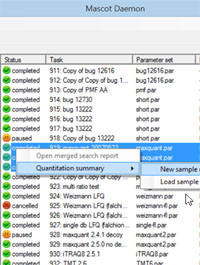
Summarizing quantitation data across multiple analyses
Quantitation studies typically include samples from different time points, treatments, fractions, and replicates, making it time-consuming (and tedious) to combine the results into a summary that can be probed for meaningful answers.
To facilitate the creation of comprehensive summaries, the latest version of Mascot Daemon allows the results of individual analyses to be selected and annotated with expression information. The results are then processed to create a spreadsheet called a Quantitation Summary, in which the rows correspond to proteins and the columns contain the expression data in the form of abundances or ratios of abundances. Both label-free and isotopic label quantitation methods are supported, including reporter ion methods.
A recent blog article illustrates the procedure using a set of 12 raw files from the label free quantitation of an E. coli digest spiked into a HeLa digest at four different concentrations. Replicates can be merged or kept separate, according to whether statistical variation across replicates is of interest. One of the strengths of the Quantitation Summary is that it uses the same rigorous protein inference as the Mascot Protein Family Summary report.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
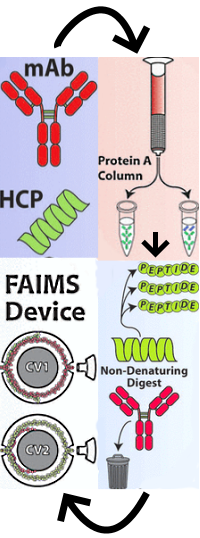
Combination of FAIMS, Protein A Depletion, and Native Digest Conditions Enables Deep Proteomic Profiling of Host Cell Proteins in Monoclonal Antibodies
Reid O'Brien Johnson, Tyler Greer, Milos Cejkov, Xiaojing Zheng, and Ning Li
Anal. Chem. 92, 10478-10484 (2020)
The authors have developed a method to identify host cell proteins (HCPs), common residual impurities in biopharmaceuticals. The method entails first depleting the sample of antibody on a protein A column, then specifically digesting HCPs while precipitating the remaining antibody under native conditions. By selectively digesting lower abundance HCPs and leaving the relatively large and stable antibody intact, they dramatically reduced the interference from antibody peptides and detected a far greater number of HCPs than in a typical digest. They then utilized high-field asymmetric waveform ion mobility spectrometry (FAIMS) to reduce the spectral congestion with LC/MS/MS.
They applied the optimized method to the NIST monoclonal antibody standard (mAb). From the combination of protein A depletion and native digestion, they detected 511 HCPs in the NIST mAb, and the addition of FAIMS expanded the sensitivity leading to an additional 20% more HCPs identified. This is several-fold more than previous studies have shown.
|
 |
 |
 |
 |
|
Mascot Tip
Several people have reported receiving an error message when they first use Database Manager after updating to Mascot Server 2.7: "Some of the predefined definitions files in /mascot/config/db_manager/public have the same last-modified time."
This is our fault. The update installs a base definition file (databases_1.xml) with the same timestamp as one of the update files that Database Manager has already downloaded, so that it is unclear which is the most recent.
Inspect a file listing to verify that this is the problem, and that the date of databases_1.xml is 8 March 2018. If so, the fix is simply to change the last modified time of databases_1.xml by a small amount. For Linux, at a shell prompt, change to the directory specified in the error message, and enter:
touch -t 201803081128 databases_1.xml
For Windows, open a PowerShell window, change to the directory specified in the error message, and enter the following text, all on a single line:
$(Get-Item "databases_1.xml").lastwritetime=(Get-Item "databases_1.xml").LastWriteTime.AddMinutes(1)
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|