|
To view this email as a web page, click here. |
 |
|
Welcome
Happy New Year!
We have the results and winners for the de novo sequencing competition from last month.
This month's highlighted publication shows a new device for automating the preparation of plasma for clinical proteomics.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns crosslink modification definitions in Unimod.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
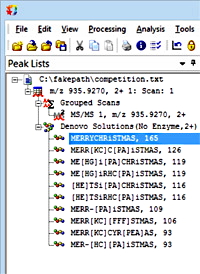
De novo competition results
The peptide sequence was MERRYCHRISTMAS, where the N-terminus and the Cys were carbamidomethylated and the Met at position 12 was oxidised. We also accepted GM or MG at the N-terminus and L instead of I at position 9, since these give identical mass values even if not so perfectly festive.
If you tried to identify the peptide by database search, you will have been disappointed. The sequence is unique to a protein from one individual, and this is not in any database because the genome has never been sequenced.
Mascot Distiller will get you most of the way if you follow the general approach described in our October 2020 blog article. The de novo algorithm is very reluctant to report solutions with more than one variable modification, so you have to cheat a bit by tightening up the fragment tolerance and make the carbamidomethyl mods fixed to get the exact solution shown here.
The first three names drawn from all the correct solutions received by 24 December were:
- Gianfranco Mamone, ISA-CNR, Italy
- Adam Dowle, University of York, UK
- Guillaume Gabant, CBM-CNRS, France
On their behalf, we have donated $300 to the WHO COVID-19 Response Fund. Kudos to everyone who submitted a correct solution. We should also acknowledge that the idea for this puzzle came from the recent EUPA/LBMSDG competition, which is referenced in the blog article.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
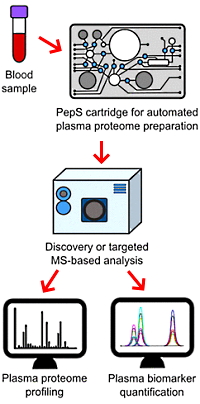
PepS: An Innovative Microfluidic Device for Bedside Whole Blood Processing before Plasma Proteomics Analyses
Benoit Gilquin, Myriam Cubizolles, Remco Den Dulk, Frederic Revol-Cavalier, Manuel Alessio, Charles-Elie Goujon, Camille Echampard, Gorka Arrizabalaga, Annie Adrait, Mathilde Louwagie, Patricia Laurent, Fabrice P. Navarro, Yohann Coute, Marie-Line Cosnier, and Virginie Brun
Analytical Chemistry published online December 15, 2020
The authors have addressed the sample preparation challenge in clinical proteomics with their development of a microfluidic device that merges several steps into an automated 2-hour process. The device encompasses plasma separation and calibration, spiking with quantification standards, albumin depletion, enzymatic digestion with trypsin, and stabilization of tryptic peptides on solid-phase extraction sorbent.
They evaluated the performance of the new device for plasma from whole blood in the context of discovery proteomics and targeted quantitative proteomics. 579 proteins were detected using the PepS cartridge compared with 535 by manual processing. Of these, 459 were detected by both methods. Among the proteins quantified, 406 (70%) were detected in all the three technical replicates when prepared with PepS compared with 429 (80%) for the manual preparations.
Analyzing a panel of three protein biomarkers routinely assayed in clinical laboratories (alanine aminotransferase 1, C-reactive protein, and myoglobin), they were able to reproducibly quantify these markers, demonstrating the potential for detecting them over their entire pathological ranges.
|
 |
 |
 |
 |
|
Mascot Tip
In Unimod, cross-links are named with the prefix Xlink:, e.g. Xlink:DST. Some of these are new-style master entries that contain information for the intact link together with expected monolinks or cleavage products. In Mascot Server 2.7, you'll find these entries listed under Linkers in the Configuration Editor.
There are also legacy entries in Unimod for individual monolinks, cleavage products and intact cross-links that happen to be intra to a single peptide. These entries can be treated as simple modifications and, in all versions of Mascot Server, you'll find these entries listed under Modifications in the Configuration Editor.
Both types of entry use the prefix Xlink: for the names. The important difference is in the classification. A specificity classified as a cross-link is a master entry that is handled by specialised code and used to link two peptides. Any other classification indicates a standard modification. More detailed information can be found on the relevant Unimod help page.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|