|
To view this email as a web page, click here. |
 |
|
Welcome
We take a look at the improvements in the latest NIST Human HCD Spectral Library.
This month's highlighted publication offers a new approach to simplify and quantify the complex methylation of histones.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month clarifies some enzyme definitions.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
NIST Human Spectral Library Update
With Mascot 2.6 and later you can readily search spectral libraries along with FASTA sequence databases to give an integrated report. You can also easily generate spectral libraries from your own search results.
The NIST Human HCD library has been updated recently, and we investigated this new version to assess its performance improvements. The original was a consensus library derived from over 10,000 raw data files, while the newer version contains 86% more peptides. Additionally, the library has now been split into three separate parts.
- human_hcd_tryp_best - high-quality spectra, mostly tryptic peptides without missed cleavages
- human_hcd_tryp_good - medium-quality spectra, mostly tryptic peptides with missed cleavages
- human_hcd_semitryp - high and medium quality spectra, mostly semi-tryptic peptides
We conducted searches of the iPRG2016 dataset against the 2016 NIST_Human_HCD, separately against each of the newer libraries, and finally a combined search against all three of the updated libraries. Searching just the updated 'best' library gave us about 20% more PSMs, but only 8 new sequences over the 2016 library. By searching all three of the updated libraries, we gained about 75% more PSMs and 324 peptide sequences.
Go here to read more details about this study and how to update your local copies of these libraries.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
A Chemical Acetylation based Mass Spectrometry Platform for Histone Methylation Profiling
Francesca Zappacosta, Craig D. Wagner, Anthony Della Pietra, III, Sarah V. Gerhart, Kathryn Keenan, Susan Korenchuck, Chad J. Quinn, Olena Barbash, Michael T. McCabe, Roland S. Annan
Molecular & Cellular Proteomics Published online March 25, 2021
The authors developed a new approach to identify and quantify the methylation of highly modified histone proteins. Since acetylation and methylation are the most common PTM's they focused on reducing the complexity due to the acetyl PTM's.
The method uses nondeuterated acetic anhydride to fully acetylate all free lysine residues prior to digestion with trypsin and analysis by LC-MS/MS. In vivo acetylation being indistinguishable from in vitro acetylation, this causes the collapse of the many in vivo acetylated forms of a given histone methyl mark into a single chemical species.
The benefit of this is demonstrated with arginine methylation of the histone H4 (1-20) peptide where acetylation has reduced the number of differently modified forms from the 68 into 6 fully acetylated species.
The authors showed selectivity and sensitivity in quantifying changes in symmetric and asymmetric dimethylation where the levels of dimethyl arginine were as low as 0.02% of the overall H4R3 occupancy. They also developed an assay to profile H3K27 methylation and applied it to an EZH2 mutant xenograft model following small molecule inhibition of the EZH2 H3K27 methyltransferase.
|
 |
 |
 |
 |
|
Mascot Tip
The naming of a couple of the standard enzyme definitions in Mascot Server can be confusing. The enzyme called None actually corresponds to non-specific cleavage. You might use this for a study of endogenous peptides. If you are doing top-down analysis of intact proteins, and don't want to simulate any type of cleavage, you need to select NoCleave.
semiTrypsin means that Mascot will search for peptides that show tryptic specificity (KR not P) at one terminus, but where the other terminus may be a non-tryptic cleavage. This is a half-way house between choosing Trypsin and None. It will only fail to find peptides that are non-specific at both ends.
(On the free public Mascot Server, None is not available for guest users because no-enzyme searches take 100 times longer than searches with tryptic specificity. In most cases, an error tolerant search will do a much better job of picking up non-specific cleavage products. If you are working with samples that are not the result of digestion with a known enzyme, and need to evaluate Mascot with such data, please email support@matrixscience.com to make a special arrangement.)
If you are using CNBr (cyanogen bromide) as a cleavage agent, remember that this converts the terminal methionine to homoserine, which may then cyclise to the lactone, so you also need to specify homoserine (Met->Hse) and/or homoserine lactone (Met->Hsl) as variable modifications. More details here.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
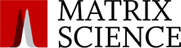 |
 |
|
|