|
To view this email as a web page, click here. |
 |
|
Welcome
Unipept (from U. Ghent) can help you analyse species and organism information in your metaproteomics search results.
This month's highlighted publication examines a range of methods for analyzing extracellular matrix proteins.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
We are also pleased to announce a new release of Mascot Distiller.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Metaproteomics with Mascot and Unipept
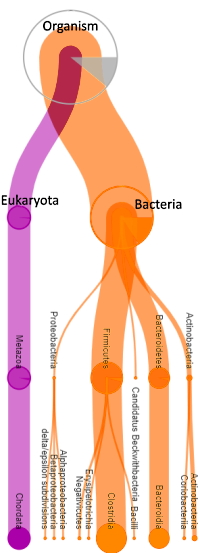
For complex samples such as gut microbes, ground water, or environmental samples, the Mascot Protein Family Summary report is a helpful tool as proteins are pulled together based on shared peptide evidence, then differentiated into family members. However, the algorithm isn't aware of taxonomy, so you will see homologous proteins from different species grouped together.
Attaching species and organism identifiers is possible by exporting Mascot results into the Unipept application. Unipept is a database created by Ghent University that maps every tryptic peptide in UniProtKB to its lowest common ancestor (LCA), which could be a single species or, if not unique to one species, a higher taxonomic rank.
The process is simple and involves a few steps of exporting the Protein Family Summary, sorting and editing some rows, and then copying the peptide sequences into a text file. This file is then imported into the Unipept online application, and results are provided in a couple minutes. The application essentially replaces conventional protein inference with LCA (e.g. species) mapping.
Go here to read more details about generating taxonomic information for your datasets.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Evaluation and refinement of sample preparation methods for extracellular matrix proteome coverage
Maxwell C. McCabe, Lauren R. Schmitt, Ryan C. Hill, Monika Dzieciatkowska, Mark Maslanka, Willeke F. Daamen, Toin H. van Kuppevelt, Danique J. Hof, Kirk C. Hansen
Molecular & Cellular Proteomics Published online April 9, 2021
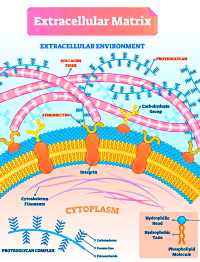
The extracellular matrix (ECM), which provides structural scaffolding and mediates signaling, has proven to be a challenge for proteomics investigators due to the poor solubility, extensive modifications, and difficult extraction of its proteins.
In this study, the authors assessed decellularization methods for their ability to extract cellular proteins while leaving ECM proteins behind using a whole mouse sample as well as four additional organs. They investigated five widely-used decellularization methods, four ECM extraction methods, and four methods for single-shot extraction and analysis. Additionally, they optimized hydroxylamine digestion methods looking at 14 sets of conditions.
They found that for optimal ECM coverage, the "Gnd-HCl/HA" extraction protocol is recommended as it produces the greatest number of PSMs and highest sequence coverage of core matrisome proteins. However, they concluded that the choice of decellularization method should be based on the objectives/priorities of a given project such as deeper proteome coverage, or focus on collagen or proteoglycans.
|
 |
 |
 |
 |
|
Mascot Distiller 2.8
We are pleased to announce the release of Mascot Distiller 2.8. Important new features include:
- Support for Bruker timsTOF data
- Ion mobility filtering in precursor quantitation for Bruker timsTOF data (requires Mascot Server 2.7)
- Global time alignment for label free quantitation
- Improved reporting using Python scripts
This free update can be downloaded from the support page.
If you wish to evaluate Mascot Distiller, send an email request to support@matrixscience.com for a product key that can be registered on-line to obtain a 30 day licence. Without a licence, Mascot Distiller operates in view-only mode.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|