|
To view this email as a web page, click here. |
 |
|
Welcome
We hope to see many of you at the upcoming ASMS meeting in Philadelphia at the end of the month. We will be present at exhibit booth #327.
Improved search speeds in Mascot Server 2.8 are the result of optimizing disk access.
This month's highlighted publication demonstrates a new set of antibodies for detecting N-terminal ubiquitination.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains when to add a new modification locally and when to add it to the public Unimod database.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Faster searches by reducing disk bottlenecks
Overall search time with Mascot 2.8 is 20-35% less than with Mascot 2.7 due to reduced disk access bottlenecks throughout the search. Running a search with 683,905 queries against the human proteome on a typical 4-core Intel Core i7 system with a traditional consumer grade hard disk, the search took 24 minutes running 2.7, while 2.8 did this in 17.5 minutes. For even more gains, a mid-grade solid state drive will be an additional ~20% faster.
As the size of data sets keeps increasing, disk access becomes more of a problem. Whether the singly threaded parts are a bottleneck depends on many factors, such as the relative time spent between the singly and multithreaded parts and the relative speed of the disk compared to the CPU. A common solution to this is to divide the computations among multiple threads and ensure the CPU isn't sitting idle.
The design we chose for Mascot 2.8 has the optimum number of processing threads to ensure that data is read and written sequentially at the maximum disk throughput. It utilizes one dedicated thread reading from disk, up to 4 threads formatting the data items, and one thread writing to disk.
Go here to read more about optimized disk speed and whether to switch to SSDs.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Antibody toolkit reveals N-terminally ubiquitinated substrates of UBE2W
Christopher W. Davies, Simon E. Vidal, Lilian Phu, Jawahar Sudhamsu, Trent B. Hinkle, Scott Chan Rosenberg, Frances-Rose Schumacher, Yi Jimmy Zeng, Carsten Schwerdtfeger, Andrew S. Peterson, Jennie R. Lill, Christopher M. Rose, Andrey S. Shaw, Ingrid E. Wertz, Donald S. Kirkpatrick & James T. Koerber
Nat. Commun. 2021, 12, 4608
The authors investigated approaches to globally profile N-terminally ubiquitinated proteins in order to better elucidate the physiological consequences of this modification. They were able to develop an antibody toolset capable of specifically detecting and enriching the tryptic remnants unique to N-terminal ubiquitination.
Using a rabbit immune phage strategy, they discovered four mAbs that selectively recognize peptides with an N-terminal diglycine-motif (GGX), but not the branched diglycine-remnant (K-ε-GG) generated by trypsin digestion of ubiquitin conjugated lysines. Lysates from HEK293 cells were digested with trypsin, and immunoaffinity enrichment was performed for GGX peptides using each of the four anti-GGX mAbs individually, with anti-K-ε-GG mAb as a control. LC/MS confirmed that the anti-GGX mAbs selectively enrich GGX peptides from cell lysates while not detecting the K-ε-GG peptides.
Using both label-free and tandem mass tag quantification they examined overexpressed cells as well as controls and were able to identify 73 substrates of the ubiquitin conjugating enzyme UBE2W.
|
 |
 |
 |
 |
|
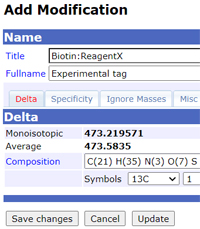
Mascot Tip
The Unimod database contains a comprehensive list of modifications, but there may be times when you need something that isn't included. Maybe an additional specificity for an existing modification or possibly an entirely new composition. It's not a good idea to add a novel modification to the public database at a very early stage. One reason is that there can be some delay between adding a new modification and seeing it in the downloadable files, which are only created when a reasonable number of changes have accumulated. Another reason is that new entries in the public database require a link to a literature reference or data sheet.
Instead, use the configuration editor of your in-house Mascot Server to add the new entry or edit an existing entry in your local copy of Unimod. It's easy to make a mistake counting up the atoms for a large modification, such as a biotin label, and local changes can be adjusted easily and immediately.
Provided you are using Mascot Server 2.5 or later, local additions and edits are not lost when you update Unimod. Once you have experimental evidence for the new modification and it has been described in a publication, then it is time to add it to the public database, making it available to other researchers.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|