|
Mass spectrometry identification of biomarkers in extracellular vesicles from Plasmodium vivax liver hypnozoite infections
Melisa Gualdrón-López, Miriam Díaz-Varela, Gigliola Zanghi, Iris Aparici-Herraiz, Ryan W.J. Steel, Carola Schäfer, Pol Cuscó, Vorada Chuenchob, Niwat Kangwangransan, Zachary P. Billman, Tayla M. Olsen, Juan R. González, Wanlapa Roobsoong, Jetsumon Sattabongkot, Sean C. Murphy, Sebastian A. Mikolajczak, Eva Borràs, Eduard Sabidó, Carmen Fernandez-Becerra, Erika L. Flannery, Stefan H.I. Kappe, Hernando A. del Portillo
Molecular & Cellular Proteomics, August 24, 2022, in press
The authors searched for potential protein biomarkers that can indicate the presence of liver hypnozoites, which are dormant malaria parasites that can reactivate and cause relapses of malarial disease. Their approach was to generate in vivo infections of P. vivax and explore the protein content of extracellular vesicles (EVs) derived from hypnozoite-infected liver cells.
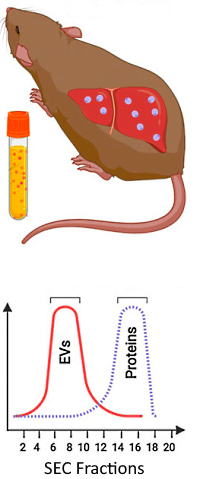
Five different groups of mice engrafted with human hepatocytes were used in these studies: uninfected control animals, mice infected with P. vivax by mosquito bite, mice infected by intravenous injection and further treated with MMV048 (to stop interfering reproduction), and those treated with the antimalarial Tafenoquine.
Plasma-derived EVs were purified by size exclusion chromatography, digested, and analyzed by LC/MS/MS using DDA. The data were searched against a customized database including P. vivax, Swiss-Prot human and mouse databases. Protein relative quantification was performed using the Abundance Estimated per sample (AEpS), which is calculated as the average areas of the top 3 most abundant peptides per protein.
The enriched SEC fractions contained proteins from the three species: 159 human, 331 mouse and 66 P. vivax. Screening of hypnozoites-specific proteins associated to EVs was done by comparing the protein ratios of P. vivax proteins across the different experimental groups, showing the presence of 13 potential candidate proteins.
|
 |