|
To view this email as a web page, click here. |
 |
|
Welcome
Can you trust your database search engine? Start by reviewing your search results.
This month's highlighted publication explores the changes in the host cell phosphoproteome upon infection with a positive-strand RNA virus.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month discusses virtualisation.
And we will be exhibiting at the upcoming HUPO 2022 Congress in Cancún, Mexico next month. Please stop by booth #411 to say hello and learn about the latest developments with Mascot products.

Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
How to trust your search engine
When you look at your database search results, a number of questions may arise: Why did the search engine identify that protein? What is the peptide evidence? Which alternatives did the software consider? What is the algorithm actually doing? In software-driven data analysis, the confidence you have in your results often comes down to trust, which has to be earned.
It is important to do spot checks and look at the peptide evidence from the bottom up, rather than assuming the computer is always right. Here are a few suggestions to evaluate your results.
You can inspect the peak matching results for every peptide match in Peptide View. Mascot records the top 10 best matches for each spectrum. Fragmentation evidence may point to sources of systematic error or poor peak picking, which can result in poor identification rates.
Statistical models rely on assumptions that could be wrong. We recommend running a target-decoy search to calibrate the error rate, which adjusts for incorrect assumptions. Compare the fragment mass evidence between target and decoy matches to check whether false discovery rate (FDR) estimation has set the score threshold to a reasonable value.
In the Protein Family Summary report, all top-level proteins are supported by at least one significant, unique peptide match. By default, Mascot displays one-hit wonders, proteins identified from a single peptide match. You can always change the format control to require two, three or more peptides per protein.
Please have a look at our blog for more suggestions.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
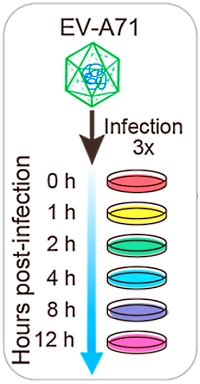
Temporal Proteomic and Phosphoproteomic Analysis of EV-A71-Infected Human Cells
Yue Zhao, Lin Li, Xinhui Wang, Sudan He, Weifeng Shi, and She Chen
J. Proteome Res., 2022, 21, 2367–2384
The authors explored the changes in the whole cell proteome and phosphoproteome upon Enterovirus A71 (EV-A71) infection of human rhabdomyosarcoma (RD) cells. The EV-A71 infects infants and children and causes hand, foot and mouth disease.
The RD cells were infected and harvested at 0, 1, 2, 4, 8 and 12h postinfection. The cells were lysed and digested, and the peptides labelled with TMT 6-plex. The labeled peptide mixture was then fractionated using three methods: strong-cation exchange (SCX) fractionation, high pH reversed-phase fractionation and regular reversed-phase column fractionation, yielding 10, 8 and 13 fractions, respectively. For the phosphoproteomic study, peptides fractionated by the regular reversed-phase column were enriched with Ti beads.
Peptides were measured using LC-MS3, which allowed quantifying 7,821 proteins with 75% quantified by at least 2 peptides. The authors also quantified more than 17,000 phosphosites, among which 10,121 were classified as high-confidence class I phosphosites that could be assigned to 3,433 proteins. They also found that the host cell phosphoproteome was changed much more than the whole cell proteome over the time course of the infection.
The authors identified several key phosphorylation signaling cascades involving kinases, which led to altered host cell functions such as protein translation, cell cycle control and cell survival, suggesting potential targets for antiviral drug development.
|
 |
 |
 |
 |
|
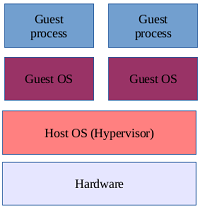
Virtualisation
We are often asked whether Mascot Server supports virtualisation and if so, how should you configure the virtual machine (VM) for optimal performance. In general, there is no issue running Mascot Server, or indeed Mascot Distiller, in a virtualised environment, but achieving good performance requires careful resource allocation.
The first and most important factor is processor virtualisation. The bare metal hardware should obviously have at least as many cores as your Mascot Server licence. If the host runs multiple VMs, you should dedicate the right number of vCPUs to the Mascot VM. Any other VM that uses the same underlying cores (vCPUs) as Mascot will without doubt slow things down. If the host processor supports hyperthreading, the hyperthreads often appear as "independent" vCPUs even though they are not equivalent to a physical core. For best performance, try to allocate only physical cores to the Mascot VM.
For random-access memory (RAM), allocate a fixed amount that is not shared with any other VM, at least 16GB and preferably 64GB. With disk space, usually the more the better. If the host runs multiple VMs, a RAID array can be beneficial to disk access, as it allows multiple VMs to read and write to the disk simultaneously.
More can be found in the Hardware virtualisation help page.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|