|
To view this email as a web page, click here. |
 |
|
Welcome to issue #105
Boost your search speed by optimizing the size of input chunks.
This month's highlighted publication shows how efficient ion mobility separations can elucidate the modification sites on histones.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot Distiller 2.8.4 has been released – update now for free.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
 |
 |
|
Speed up your search with this 'one little trick'
When you submit peak lists for a database search, Mascot sorts the spectra by peptide mass and splits them into chunks. This reduces peak RAM usage during the search and provides very efficient memory caching. Chunking also provides effective load balancing in cluster mode, because the search space is identical on each node.
The default chunk size is 1000 queries (MS/MS spectra), but increasing this to 2000 can yield an immediate 10-20% performance boost, at the expense of higher RAM usage. The parameter is called SplitNumberOfQueries and is complemented by another parameter, SplitDataFileSize, which limits the size of the chunks in bytes.
The various hardware, software and data combinations can have a large effect on the optimum value for these parameters, so the only way to find your best settings is to run a set of benchmarking tests.
Go to our blog for a suitable procedure, using a publicly available multi-species data set.
We tested a mid-range Intel Core i5 laptop as well as a workstation with a single AMD Ryzen 9 processor with Mascot installed on a RAID array. Increasing the SplitNumberOfQueries to 1500 on the laptop and 2000 on the workstation produced 10% and 21% boost in search speed, respectively.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
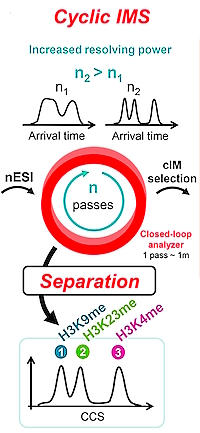
Full Separation and Sequencing of Isomeric Proteoforms in the Middle-Down Mass Range Using Cyclic Ion Mobility and Electron Capture Dissociation
Francis Berthias, Dale A. Cooper-Shepherd, Frederik H. V. Holck, James I. Langridge, and Ole N. Jensen
Anal. Chem. 95, 29, 11141–11148
The authors investigated the ability of high-resolution cyclic ion mobility (cIM) coupled with electron capture dissociation (ECD) to separate and identify PTM positional isomers of histone N-tails. They used a set of synthetic mono- and trimethylated isomers, H3K4me1/H3K9me1/H3K23me1 and H3K4me3/H3K9me3/H3K23me3.
Full separation of the mixture of monomethylated isomers is achieved for the charge state 10+, exhibiting three distinct peaks at 151.6, 153.3 and 156.5 ms, corresponding to H3K9me1, H3K23me1 and H3K4me1. The full separation of the trimethylated isomers is obtained at charge state 11+ with r = 0.90, 2.18 and 1.28 for H3K4me3/H3K9me3, H3K4me3/H3K23me3 and H3K9me3/H3K23me3, respectively.
MS/MS (ECD) spectra confirmed the cIM separation and provided 90−100% amino acid sequence coverage. Additionally, these spectra confirmed the locations of the methyl groups for the various monomethyl and trimethyl isomers.
|
|
 |
 |
 |
|
Mascot Distiller 2.8.4
Mascot Distiller 2.8.4 has been released. This minor patch release adds support for Agilent 6546 LC/QTOF, fixes a charge state issue with Thermo Orbitrap Exploris 480 and addresses a few important bugs. All users of Distiller 2.8 should install the 2.8.4 patch, which is free of charge.
If you are using an earlier version, updating to the latest version is currently free, until the next major version! Distiller 2.8 made big improvements in all areas of the software, including label-free quantitation, time alignment and quantitation reports. You do not need a new product key. Just download and install the latest version and your existing licence will work.
Download the installer from our website.
The full change history can be found in the release announcement.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot. Read more about the company on our about page.
|



|
 |
|
|