|
To view this email as a web page, click here. |
 |
|
Welcome to issue #106
When writing up a methods section, it is important to state the version of the data analysis software.
This month's highlighted publication explores a new methodology for characterizing glycoproteins.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot Server 2.8.3 has been released. Check below how to download this free update.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
 |
 |
|
How old is the software in that publication?
Think back to the most recent proteomics paper you read.
Did the methods section state the version number of the software used for protein identification?
This is essential information for assessing the quality and reproducibility of the data analysis.
As an example, if you identified endogenous peptides using Mascot Server 2.4, which was released a decade ago, and then repeated the search using version 2.8, you will find both the previous set of peptides and many more that the old version could not.
It is also helpful to state the software release date or year. Many software packages have frequent releases, and it is rarely obvious from the version number how old the software is.
The lead time from planning and running the experiment to publishing a paper can be long, and sometimes there is a good reason to stick to an old version. On the other hand, if the software used in the experiment is very old, how much could the data analysis have been improved by using a more recent version?
Go to our blog to read more about reproducibility and how to find the Mascot Server release date from its version number.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Mapping the Human Chondroitin Sulfate Glycoproteome Reveals an Unexpected Correlation Between Glycan Sulfation and Attachment Site Characteristics
Fredrik Noborn, Jonas Nilsson, Carina Sihlbom, Mahnaz Nikpour, Lena Kjellén, Göran Larson
Mol Cell Proteomics (2023) 22, 100617
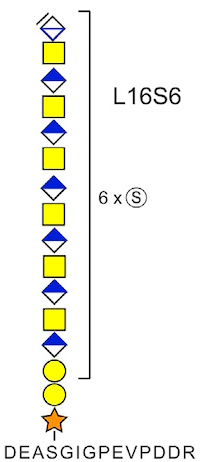
The authors have developed methods to identify the core peptides and linkages for human chondroitin sulfate proteoglycans (CSPG). These are composed of chondroitin sulfate (CS) polysaccharides attached to different core proteins.
The identification is achieved by trypsin digestion of CSPG-containing samples, followed by CS glycopeptide enrichment using strong-anion-exchange chromatography. The enriched glycopeptides are then incubated with chondroitinase ABC to depolymerize the CS polysaccharides, generating polysaccharide (6-18mer) residual structures including the linkage regions still attached to the peptides.
Samples were analyzed by LC-MS/MS, and database searching included variable modifications of the residual 6-mer structure for [HexA(-H2O)GalNAcGlcAGalGalXyl-O-] with 0, 1, or 2 sulfates or phosphates attached. This identified 21 previously established CSPGs as well as five novel CSPGs. Similar results were obtained for the 8-18mers. Statistical analysis of these results indicated that the level of sulfation correlated with the acidity of the attachment motifs.
|
|
 |
 |
 |
|
Mascot Server 2.8.3
Mascot Server 2.8.3 is now available.
This patch fixes six small bugs in the search engine, four in Mascot Perl scripts, one in ms-monitor.exe regarding a spurious log message and one issue with licensing on AMD EPYC processors.
The announcement gives the details.
All users with version 2.8 licence should install the update. The cumulative update from 2.8.0, 2.8.1 and 2.8.2 can be
downloaded from the support page (scroll down to Windows Update Procedure or Linux Update Procedure).
The cumulative list of changes since 2.8.0 is shown at the bottom of the page.
If your support contract has run out and you would like to renew it, please contact us. Patch 2.8.3 is free to download and install, but staying under support means you will receive future software updates for free.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot. Read more about the company on our about page.
|



|
 |
|
|