|
To view this email as a web page, click here. |
 |
|
Welcome to issue #108
Mascot can identify peptides in chimeric spectra from complex mixtures or wider isolation windows.
This month's highlighted publication demonstrates a method to characterize integral membrane proteins.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month is the patch release of Mascot Distiller 2.8.5.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
 |
 |
|
Identifying peptides from chimeric spectra
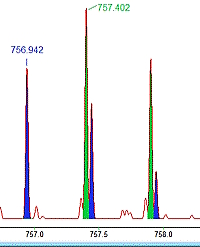
If you analyze complex mixtures like environmental samples or use a broad precursor isolation window as in DIA, then it is likely that multiple precursors are selected for fragmentation at the same time, producing chimeric spectra. This presents a challenge for peptide identification by database searching.
Mascot Server has supported chimeric spectrum searching for almost 10 years. In a simple case, if you know that multiple precursors were fragmented into the same spectrum, you can just add another PEPMASS line in the Mascot Generic Format (MGF) file. And if your peak picking software doesn't produce MGF files with multiple PEPMASS lines per peak list, give Mascot Distiller a go.
When Mascot reads the peak list, it is split into two (or more) subsidiary queries, one per PEPMASS line, where each query gets a copy of the original peak list. During the database search, peptides are matched and scored to each query independently. Mascot treats the unknown fragment peaks from the other precursor as unexplained noise. This approach has many advantages. For example, the peptide match has to have unambiguous fragmentation evidence to get a decent score.
It's not always easy to find and navigate chimeric matches. We've created a basic chimericity report script that you can download and use with any version of Mascot. The script summarizes the number of precursors per spectrum in the peak lists, as well as provides a breakdown of the number of significant peptides matches to spectra of increasing chimericity. Additionally, to get a better FDR estimate, we recommend changing the decoy type from reverse to random.
Please go to our blog to read more about utilizing Mascot Server for searching chimeric spectra.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Liquid-chromatography mass spectrometry describes post-translational modification of Shewanella outer membrane proteins
Jessica H. van Wonderen, Jason C. Crack, Marcus J. Edwards, Thomas A. Clarke, Gerhard Saalbach, Carlo Martins, Julea N. Butt
BBA - Biomembranes 1866 (2024) 184221
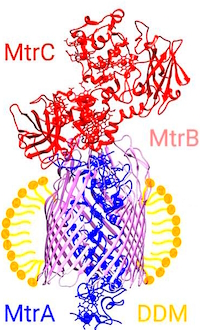
The authors have developed an LC/MS method to investigate the three protein members of the integral membrane complex, MtrCAB, purified from Shewanella bacteria. The method utilizes a two-step protocol for complete removal of detergents by application of aqueous methanol while the proteins remain tightly bound to the column. Proteins are subsequently resolved by gradient elution with aqueous acetonitrile and directly infused into the MS. This allows the membrane proteins, initially suspended in the non-ionic detergent dodecyl maltoside (DDM), to be cleanly separated from detergent prior to protein elution.
The intact mass values for the MtrA and MtrB proteins matched their predicted values (38,570, and 75,052, resp.) within 2Da, while the MtrC protein was measured at 552Da more than expected. MS/MS yielded good coverage of MtrA (64%), MtrB (94%) and MtrC (74%) but none of the MtrC peptides provided evidence for PTMs suggested by the intact mass analysis. An engineered form of MtrC (without terminal PTM) did match the predicted mass. Coupled with other work showing lipid PTMs, they concluded that the MtrC protein identified in this work is most reasonably the result of diacylation by palmitoleic acid of the N-terminal cysteine.
|
|
 |
 |
 |
|
Mascot Distiller 2.8.5
We have recently released Mascot Distiller 2.8.5. This patch release includes bug fixes to hierarchical clustering reports, tooltip text, sequence tags and PMF searches.
We recommend that all users download and install it.
Follow the link on the Distiller product page for the download link.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot. Read more about the company on our about page.
|




|
 |
|
|