|
To view this email as a web page, click here. |
 |
|
Welcome to issue #109
The ABRF iPRG 2023 crosslinking study runs until 15 January – act now if you want to participate! Use the free Mascot service or your in-house licence to analyse the data.
This month's highlighted publication utilizes CRISPR knockouts to show the role of N-terminal acetylation on protein stability.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Enable internal fragments in Mascot instrument settings to get better results.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
 |
 |
|
ABRF iPRG 2023 Crosslinking Study
The ABRF proteome informatics research group (iPRG) has launched a new crosslinking study:
The goal of this study is to gain information from the proteomics community about the different approaches for crosslinked peptide MS data analysis, showcasing the multitude of different pipelines and tools available, and providing guidance for improving the presentation of crosslinked MS results.
We have a Mascot tutorial to help you obtain the best results from your analysis. It's a walk-through of the required steps:
- Prepare and configure Mascot Server for this data set
- Process the data automatically with Mascot Server and Distiller
- Evaluate the crosslinking results
- Export the results to xiVIEW for further analysis and visualization
If you don't have your own Mascot Server and would like to try searching this data set on the public Mascot Server, you are welcome to do so. However, please contact us to request a free temporary account with expanded limits so you can search the full data set.
The data is high resolution MS/MS, and we processed it with Mascot Distiller. Crosslinked peptides can have quite high masses, which means they often have higher charge states too. We recommend either decharging the fragment ions to MH+ or exporting their charge state in the MGF file, both of which help a lot in the database search.
The final part of the tutorial covers analysis of the results. We have previously published some guidelines for validating intact crosslinked peptide matches, and you can use those guidelines to determine what a good match looks like and which ones don't have sufficient information to validate the crosslink.
Go to our blog to download the full tutorial and data files.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
N-terminal acetylation shields proteins from degradation and promotes age-dependent motility and longevity
Sylvia Varland, Rui Duarte Silva, Ine Kjosås, Alexandra Faustino, Annelies Bogaert, Maximilian Billmann, Hadi Boukhatmi, Barbara Kellen, Michael Costanzo, Adrian Drazic, Camilla Osberg, Katherine Chan, Xiang Zhang, Amy Hin Yan Tong, Simonetta Andreazza, Juliette J. Lee, Lyudmila Nedyalkova, Matej Ušaj, Alexander J. Whitworth, Brenda J. Andrews, Jason Moffat, Chad L. Myers, Kris Gevaert, Charles Boone, Rui Gonçalo Martinho & Thomas Arnesen
Nat Commun 14, 6774 (2023)
The authors investigated the role of N-terminal acetylation in protein stability and degradation, as well as its impact on cellular processes, longevity, and motility. The study used genome-wide CRISPR knockout screens in human HAP1 cells and fruit fly Drosophila melanogaster. They determined that N-terminal-acetylation protects hydrophobic N-termini against targeted degradation and that acetyltransferase NatC perturbation appears to regulate vesicle trafficking and organelle morphology.
The study utilized next-generation sequencing, flow cytometry analysis, cell sorting, confocal imaging, and immunoblot analysis as well as MS proteomics. For the MS analysis, they enriched N-terminal peptides by using low pH strong cation exchange chromatography. To enable the assignment of in vivo N-terminal acetylation events, all primary protein amines were blocked at the protein level with N-hydroxysuccinimide ester of acetic acid encoded with heavy isotopes. This made it possible to distinguish between in vivo and in vitro acetylated N-termini and to calculate the degree of in vivo Nt-acetylation. Spectra were searched twice, one search to quantify the N-terminal acetylated peptides and another search to identify all the peptides including the ones that are not N-terminally acetylated. The degree of in vivo acetylation was calculated separately using the light/heavy ratio reported by the Mascot Distiller Quantitation Toolbox.
Label-free quantitation on HAP1 WT and acetyltransferase KO cells identified 440 proteins that were differentially expressed between WT and KOs using multiple ANOVA testing. 346 of the ANOVA significant proteins were also significant in at least one pairwise comparison, of which 66 % were depleted while 34% were enriched compared to WT.
|
|
 |
 |
 |
|
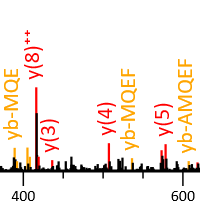
Get better HCD results by matching internal fragments
Higher-energy collisional dissociation (HCD) tends to produce internal cleavage ions, especially with stepped collision energy. Mascot supports matching internal fragments, but the feature is not enabled by default. We recommend trying it if you use HCD.
Our July 2023 blog has a very nice example provided by Darryl Pappin, Cold Spring Harbour Laboratory. It's a QC injection of 81,267 queries, and simply enabling the ya and yb series gives 16% more matches at 1% PSM FDR and 13% more peptide sequences. Enabling these series allows Mascot to label internal fragment peaks as internals, rather than noise, which tends to improve the match score. It's also beneficial in crosslinking studies, where overall fragmentation is often sparse, and sometimes internals provide the only good evidence for one or the other of the two crosslinked chains.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot. Read more about the company on our about page.
|




|
 |
|
|