|
To view this email as a web page, click here. |
 |
|
Happy New Year!
Narrow window DIA datasets can be readily analyzed using Mascot with a spectrum-centric approach.
This month's highlighted publication shows the measurement of ubiquitylated proteins involved in DNA damage repair.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Windows Server 2019 mainstream support is ending – Mascot Server 2.7 and earlier are affected.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
 |
 |
|
Spectrum-centric searching of narrow window DIA data
Data Independent Acquisition (DIA) spectra are most commonly analysed with a peptide-centric approach, where a library of experimental or predicted MS/MS spectra are compared to the observed spectra.
In contrast, spectrum-centric DIA utilizes a standard database search of uninterpreted MS/MS data, avoiding the requirement for either experimental or predicted spectral library.
This approach has clear benefits in handling unexpected modifications – with an experimental library the match needs to be present, and for a predicted library approach the modification of interest needs to have been included in the training data used for the model.
A spectrum-centric match is easier to interpret and visualize as each identification has to stand on its own merits. Also, because the spectra are searched independently, the search scales easily across multiple processors.
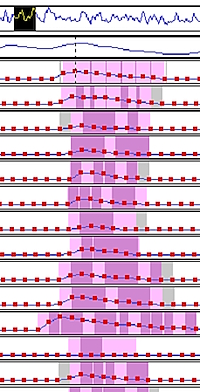
We used Mascot Server and Mascot Distiller to analyse a narrow window (8 m/z) DIA dataset from a prostate cancer study, which looked at 20 cancer and non-cancer patients. Peak picking and search submission were automated with Mascot Daemon. At the selected 1% PSM FDR, we found ~1.6 million significant PSMs, covering 4296 protein hits and ~20.5 thousand peptide sequences.
We also ran label-free quantitation on the entire dataset using time alignment and match between runs. Because we’re taking a spectrum-centric approach, we’re seeing peptide matches across the entire XIC region for most components, which gives a high degree of confidence in the identifications.
Go to our blog to read more about narrow window DIA.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Dissecting ubiquitylation and DNA damage response pathways in the yeast Saccharomyces cerevisiae using a proteome-wide approach
Ewa Blaszczak, Emeline Pasquier, Gaëlle Le Dez, Adrian Odrzywolski, Natalia Lazarewicz, Audrey Brossard, Emilia Fornal, Piotr Moskalek, Robert Wysocki, Gwenaël Rabut
Molecular & Cellular Proteomics, Published online: December 13, 2023
Ubiquitin plays a critical role in DNA damage response (DDR) by regulating the localization, activity and stability of DDR-associated proteins. Failure to repair DNA damage is the underlying cause of cancer development and premature aging in humans. In this study, the authors investigated the extent of ubiquitylation in yeast that is treated with the DNA alkylating agent methyl methanesulfonate (MMS).
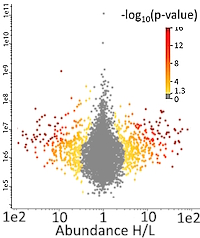
Yeast cells were grown in a medium (without lysine and leucine) supplemented with either "light" (12C614N2) or "heavy" (13C614N2) lysine. In the last hour of incubation, MMS was added to cells cultured with "heavy" lysine. Cells cultured with and without MMS were mixed in 1:1 ratio, harvested, and the proteins extracted. Ubiquitylated proteins were enriched by immobilized metal affinity chromatography, then digested with Lys-C followed by trypsin. Ubiquitylated peptides were further purified using anti-K-ε-GG antibody-coated beads. These peptides were fractionated on reverse phase media, and the fractions analyzed by LC/MS-MS. For database searching, besides methionine oxidation, variable PTMs were set for the ubiquitin di-glycine remnant (-GG signature, 114.04 Da), and Leu-Arg-Gly-Gly remnant (-LRGG signature generated by one missed trypsin cleavage, 383.23 Da).
The investigators quantified 5397 distinct K-ε-GG peptides corresponding to 1853 proteins. Among these, 556 peptides from 473 proteins were upregulated more than 2-fold in response to MMS treatment while 644 peptides from 513 proteins were downregulated. The peptides significantly upregulated in response to MMS enabled the localization of 519 ubiquitylated residues in 435 proteins, among which 260 (50%) are not referenced in the BioGrid interaction repository.
|
|
 |
 |
 |
|
Windows Server 2019
By the time you read this newsletter, Microsoft has ended Windows Server 2019 mainstream support, which was on 9 January 2024.
Windows 10 mainstream support will end 18 months later, in October 2025.
If you're running Mascot Server on Windows, Mascot will continue to work, but your IT people may increasingly demand that you update the operating system.
The good news is, Mascot Server 2.8 fully supports Windows 11 and Server 2022.
Updating Windows is straightforward and won't break Mascot.
However, Mascot Server 2.6 and 2.7 are incompatible with Windows 11 and Server 2022 because of a fault in a third-party Perl module shipped with Mascot.
Either Database Manager stops working or all Mascot CGI scripts stop working, depending on which Windows updates have been installed.
We have also seen this issue on some Windows Server 2019 systems after the October 2023 Windows security update (OS Build 17763.4974).
The easiest and quickest solution is to update to Mascot Server 2.8.
Please contact us for a quote and migration instructions.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot. Read more about the company on our about page.
|




|
 |
|
|