|
To view this email as a web page, click here. |
 |
|
Welcome
Are you studying an organism with an unsequenced genome? If so, expressed sequence tags might be the best database for searches.
This month's highlighted publication features a method to identify proteins and their localization in specific endomembrane compartments.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month is what to do if you forget your Mascot Security password.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Searching with Expressed Sequence Tags
If the genome of your organism of interest has not been sequenced, its proteins will not be included in the major
public sequence databases, such as NCBInr or UniRef100. Although proteins from closely related strains or species may be
sufficient to get partial matches, it is well worth checking coverage in the vast databases of expressed sequence tags (ESTs).
For example the Atlantic cod has only 1,207 distinct protein sequences in NCBInr compared with 257,435 ESTs in GenBank.
To view entry counts for your organism in the various databases derived from GenBank, you can
use the NCBI taxonomy browser.
The EST databases are highly redundant and most entries are just fragments of full length proteins.
But, if you are lucky, and there is a UniGene index for your organism of interest, you can compensate for this and
obtain high quality search reports by mapping the redundant EST accessions into gene families.
More details on this procedure are shown in this online presentation
and this document.
For more information about ESTs as well as the deluge of nextgen sequence data, please
go here. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
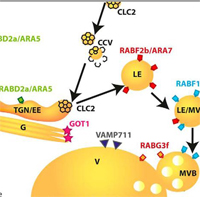
Identification of Regulatory and Cargo Proteins of Endosomal and Secretory Pathways in Arabidopsis thaliana by Proteomic Dissection.
William Heard, Jan Sklenář, Daniel F. A. Tomé, Silke Robatzek and Alexandra M. E. Jones
Molecular & Cellular Proteomics (2015) 14: 1796-1813.
This paper investigates an approach to decipher the membrane compartmentalization, through which cells separate and control biological processes. Plant growth, development, and adaptation to biotic and abiotic stress all rely on the dynamic endomembrane system, yet there is little known about the proteins regulating these dynamic trafficking events.
The authors developed a method based on affinity purification of fluorescent fusion constructs to seven subcellular marker proteins in Arabidopsis thaliana seedlings. A total of 433 proteins were presented, only five of which were shared among all enrichments, while many proteins were common between endomembrane compartments of the same trafficking route. Approximately half were assigned to only one enrichment.
More details as well as datasets are available from the proteomics database PRIDE (PXD001877 to PXD1885 inclusive).
|
 |
 |
 |
 |
|
Mascot tip of the month
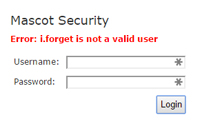
If Mascot Security is enabled on your Mascot Server and you forget your password, you'll need to ask your system administrator
to reset it. Its no good contacting Matrix Science support - passwords are set and saved locally on your server.
But, what if you are the system administrator and you forget the master password?
Fortunately, there is an easy way to reset it. Open a command window on a Windows server or a shell prompt on a Linux server. Change to the Mascot bin directory and execute the script enable_security.pl. You'll see the re-assuring message that the password for the admin account has been reset to admin. You are forced to change this to something more secure when you next log in, which you should do immediately. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|