|
To view this email as a web page, click here. |
 |
|
Welcome
As instrument accuracy gets better and better, de novo sequencing becomes a more practical proposition for peptide sequences that aren't in any database. Did you know that the Mascot Distiller Search Toolbox includes a powerful de novo algorithm? We describe how this complements database search.
Halloween's highlighted publication shows the characterization of a giant virus that is still infectious after 30,000 years frozen in permafrost.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes how to obtain intensity values for individual components in a quantitation experiment.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
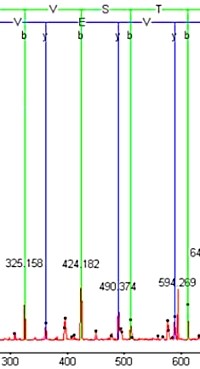
Dig deeper with de novo sequencing
Whether you are studying an organism with an unsequenced genome or spectra that cannot be identified due to an unsuspected modification or alternative splicing, de novo sequencing can help.
Current generation instruments can achieve unprecedented levels of mass accuracy and signal to noise, but this doesn't guarantee unambiguous, end-to-end peptide sequences. The peptide may be unwilling to fragment along its entire length, or the mass values may fall in ways that allow multiple interpretations. A common solution is to try to align the de novo sequence against a database sequence, using an error tolerant sequence tag, allowing attention to be focused on the novel region that contains the mutations or modifications.
The Mascot Distiller Search Toolbox incorporates a powerful de novo algorithm. This can be used in combination with Mascot Server to implement a strategy such as the following:
- Standard Mascot database search returns the easy matches.
- Error tolerant database search returns additional matches, but only for proteins already identified.
- Automated de novo sequencing of all the unmatched spectra.
- Error tolerant sequence tag search of the unambiguous regions of the de novo results.
Go here to learn more about combining database search and de novo. Note that Mascot Distiller can be evaluated using a free 30-day trial |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
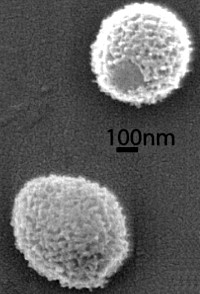
In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba
Matthieu Legendre, Audrey Lartigue, Lionel Bertaux, Sandra Jeudy, Julia Bartoli, Magali Lescot, Jean-Marie Alempic, Claire Ramus, Christophe Bruley, Karine Labadie, Lyubov Shmakova, Elizaveta Rivkina, Yohann Coute, Chantal Abergel, and Jean-Michel Claverie
PNAS (2015), vol. 112, E5327-E5335
This paper investigates an ancient virus that was discovered in the Siberian permafrost, and shown to still be infective. It is a very large spherical virion (0.6-µm diameter) enclosing a 651-kb GC-rich genome encoding 523 proteins.
The proteome of purified Mollivirus particles is associated with up to 230 proteins, each of them detected by the identification of at least two different peptides using tandem mass spectrometry. The proteome of the particle revealed two main features: the absence of an embarked transcription apparatus and the unusual presence of many ribosomal (and ribosome-related) proteins.
In addition the authors examined the variation of expressed functions and proteins occurring in A. castellanii infected by Mollivirus by performing a series of analyses at regular intervals throughout the whole virus replication cycle. A description of the workflow and the mass spectrometry data have been deposited in PRIDE - PXD002374 and PXD002375 |
 |
 |
 |
 |
|
Mascot tip of the month
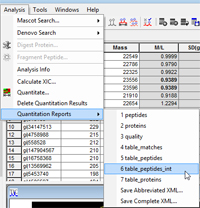
Whether you are using Mascot Distiller for MS1 quantitation or Mascot Server for MS2 quantitation, results are reported as a set of ratios. If you need intensities for individual components, these can also be obtained.
For Mascot Distiller, download this report template and unzip to a file called table_peptides_int.xslt in the Mascot Distiller reports directory (probably C:\Program Files\Matrix Science\Mascot Distiller\reports). This will automatically create a new entry on the Mascot Distiller quantitation reports menu. The new report, intended for copying and pasting to Excel, includes the start and end retention times for each XIC in addition to the integrated intensity. Note that a ratio calculated from two intensities may differ from the reported ratio unless you select 'simple ratio' in the quantitation method. This is because the default method of calculating a ratio for precursor protocol is to fit a straight line to a cross-plot of the intensity values from each spectrum in the XIC, as described in the Distiller help under 'Quick tour SILAC Example'.
For Mascot Server, intensity values are available in the configurable CSV and XML exports. Make sure you check both Protein quantitation and Peptide quantitation. For the CSV format, you'll find the peptide quantitation information in the rightmost columns: first, columns for each ratio, then for each intensity. Protein quantitation is placed to the right of the peptide data on the last row for each protein.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|