|
To view this email as a web page, click here. |
 |
|
Happy New Year!
We explain how easily you can create a customised table of confidently identified proteins with just a few mouse clicks.
This month's highlighted publication shows how a combined mammalian database can provide answers for the sea lion, which has an unsequenced genome.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes two very brief tutorials that are ideal for getting up to speed on the essentials of PMF and MS/MS searching.
If you are booking travel or a hotel for ASMS in San Antonio, please note that we will be holding breakfast meetings on Monday and Tuesday instead of a user meeting on Sunday morning.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
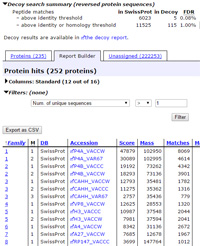
Listing confidently identified proteins
Nothing could be easier than creating a customised table of confidently identified proteins in Mascot 2.5 using Report Builder. The key steps are:
- Select the Decoy checkbox when submitting the search
- Open the result report as a Protein Family Summary
- Switch to the Report Builder tab
- Expand the decoy search section and set the peptide FDR to 1%
- Expand the filters section and set 'Num of significant unique sequences' > 1
- Optionally, expand the columns section and choose which columns you require and their order
- Print the table or export it as CSV
Step 5 removes the 'one-hit wonders'. Many other useful filters are available. For example, you can filter by database so as to remove contaminants from the final table. For more detailed information, see this blog article. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Proteomic analysis of cerebrospinal fluid in California sea lions (Zalophus californianus) with domoic acid toxicosis identifies proteins associated with neurodegeneration.
Benjamin A. Neely, Jennifer L. Soper, Frances M. D. Gulland, P. Darwin Bell, Mark Kindy, John M. Arthur, and Michael G. Janech
Proteomics (2015) 15: 4051-4063
For the past 20 years, California sea lions have suffered mass strandings thought to be caused by ingesting prey that have concentrated toxins from algae blooms. These domoic acid toxins (DAT) cause neurodegenerative disorders by crossing the brain-blood barrier and interfering with nerve signal transmission.
Diagnosis of DAT is problematic because domoic acid is rapidly cleared from the body and may not be directly detected when stranded sea lions are ultimately found. Additionally, proteomics is difficult since the genome of the sea lion is not sequenced. To overcome this, the authors searched against protein sequences from several better characterised terrestrial mammals.
From the LC/MS/MS of 11 samples, a total of 206 proteins were identified (FDR < 0.1) using the composite mammalian database. Several peptide identifications were validated using stable isotope labeled peptides. Comparison of spectral counts revealed seven proteins that were elevated in the cerebrospinal fluid from sea lions with DAT. Mass spectra have been uploaded to ProteomeXchange (dataset PXD002105). |
 |
 |
 |
 |
|
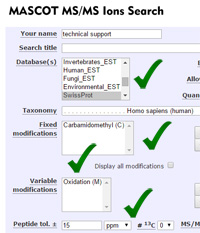
Mascot tip of the month
We try to keep the Mascot search form as simple as possible, but there is still a steep learning curve associated with database searching. Ideally, anyone new to the field would follow our webcast training course, as well as reading around the subject in the literature. The reality is that few people have the time for this.
If you are new to MS-based proteomics, or if there are new people in your group, don't miss the short tutorial articles linked from the Access Mascot Server page on our web site. Estimated reading time is just 8 minutes for the peptide mass fingerprint tutorial and 12 minutes for the one on searching MS/MS data.
We like to think these tutorials cover the essentials and, even if you read nothing else, you won't go too far wrong. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|