|
To view this email as a web page, click here. |
 |
|
Welcome
Please join us at our upcoming ASMS Breakfast Briefings on June 6th and 7th in San Antonio.
If you ever need to study selenocysteine-containing proteins, we have some suggestions.
In this month's featured publication, ribosomal profiling and proteomics identify the translation start sites of expressed genes in bacteria.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains the importance of the database *.stats file.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Join Us at ASMS in San Antonio
We would like to invite you to join us for breakfast
at the upcoming ASMS meeting in San Antonio in June. We are hosting two sessions where you can learn about the latest applications and developments
in database search and quantitation. There is no charge for attending these meetings, but advance registration is required.
CLICK HERE FOR DETAILS AND REGISTRATION
Monday 6 June, 7:00 am - 8:00 am
Ancient proteomes - proteomics in archaeology, palaeontology and forensics
presented by Michael Buckley, Manchester University
Mass tolerant and error tolerant searches: how do they compare?
presented by Matrix Science
Tuesday 7 June, 7:00 am - 8:00 am
From Samples to Biomarker Discovery: The keys for Large Scale Proteomics
presented by John Corthesy, Nestlé Institute of Health Sciences
New features in Mascot Distiller - MSE, MS3 reporter ion quant, and more
presented by Matrix Science
|
 |
 |
 |
 |
|
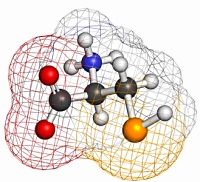
Selenocysteine and U
Selenocysteine (U) is a rare residue, but there are 46 instances of it in the human proteome and if you happen to be studying selenoprotein P, which contains 10 U residues, you definitely need to allow for the residue in both unmodified and modified forms. Since it was only recently added to Unimod as a target residue, you may not see it in your Mascot Server configuration editor. This blog article explains exactly what you need to do to update your local Unimod file.
After updating Unimod, you should find that the search form lists these modifications for selenocysteine, plus any that you have defined locally:
- Carbamidomethyl (U)
- Carboxymethyl (U)
- Dioxidation (U)
- MolybdopterinGD (U)
- Oxidation (U)
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
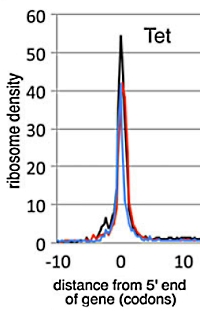
Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling
Kenji Nakahigashi, Yuki Takai, Michiko Kimura, Nozomi Abe, Toru Nakayashiki, Yuh Shiwa, Hirofumi Yoshikawa, Barry L. Wanner, Yasushi Ishihama and Hirotada Mori
DNA Res (2016) online: March 23, 2016
The precise determination of protein coding genes in bacteria has proven to be difficult, and this paper presents an additional approach to further elucidate the translation start sites of expressed genes in bacteria. The authors used tetracycline-inhibited ribosome profiling to confirm the translation sites of e. coli. The tetracycline inhibits translation by preventing the stable binding of tRNA to the ribosome.
The approach showed that start sites identified here correspond >70% of the start site changes in the latest (2014) GenBank annotation record, indicating a good level of sensitivity. They used β-galactosidase gene fusions to confirm new translation start sites, and employed proteomics to further confirm by selectively enriching N-terminal peptides after tryptic digestion and LC-MS identification of peptides.
They also discovered over 300 translation start sites within non-coding, intergenic regions of the genome, which could correspond to pseudogenes, small peptides or have unknown roles. |
 |
 |
 |
 |
|
Mascot tip of the month
For each database in Mascot, you can view useful information by following the statistics link on the Database Status page. It is important to check that the numbers look reasonable for a new database, and worth looking occasionally after updates, in case something has changed or the download was truncated. The main sections are:
- A header, with counts for the numbers of sequences and residues. There is also a count for 'invalid residues'. If this is ever non-zero, it needs investigation.
- Counts for each taxonomy category. If the database has taxonomy defined, check that it is working correctly and each category is populated
- Counts for each residue. If you have specified the database as AA and most of the residues are A, C, G, and T then you may be disappointed with the search results
- Counts of sequences by length.
- Counts of sequences by the number of associated taxonomy IDs. If your taxonomy indexes are up-to-date, every entry in Swiss-Prot should have a single taxonomy ID. For a database like NCBInr, there will usually be a few entries that have no taxonomy ID, the majority will have one, and some will have many because identical sequences from different organisms have been collapsed into a single entry. For accurate searching, make sure the fraction of entries with no taxonomy ID is well below 0.1%.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|