|
The epichaperome is an integrated chaperome network that facilitates tumour survival
Anna Rodina, Tai Wang, Pengrong Yan, Erica DaGama Gomes, Mark P. S. Dunphy, Nagavarakishore Pillarsetty, John Koren, John F. Gerecitano, Tony Taldone, Hongliang Zong, Eloisi Caldas-Lopes, Mary Alpaugh, Adriana Corben, Matthew Riolo, Brad Beattie, Christina Pressl, Radu I. Peter, Chao Xu, Robert Trondl, Hardik J. Patel, Fumiko Shimizu, Alexander Bolaender, Chenghua Yang, Palak Panchal, Mohammad F. Farooq, Sarah Kishinevsky, Shanu Modi, Oscar Lin, Feixia Chu, Sujata Patil, Hediye Erdjument-Bromage, Pat Zanzonico, Clifford Hudis, Lorenz Studer, Gail J. Roboz, Ethel Cesarman, Leandro Cerchietti, Ross Levine, Ari Melnick, Steven M. Larson, Jason S. Lewis, Monica L. Guzman & Gabriela Chiosis
Nature 538, 397-401 (20 October 2016)
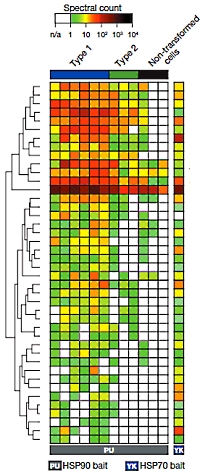
The authors have investigated the multi-protein complexes centered around the chaperones, heat shock proteins 70 and 90, and have determined that they are part of a larger network encompassing numerous cellular processes vital for tumor cell function. They term this larger network the epichaperome.
They also found that this network is active in certain types of cancer cells, giving them a degree of protection. However, disruption of the chaperone complexes causes dismantling of the network and cell death. They investigated a range of cancer cell lines (pancreatic, gastric, lung, and breast cancers, as well as lymphomas and leukaemias) and found that approximately 60-70% presented medium to high levels of epichaperome complexes. Additionally the oncogene MYC was able to rewire the network and to drive the formation of the epichaperone. The vulnerability of the network to disruption may pose opportunities for pharmaceutical interventions. |
 |