|
To view this email as a web page, click here. |
 |
|
Welcome
Please join us at our Breakfast Workshops at the upcoming ASMS Conference in Indianapolis, June 5th and 6th.
A search result is only as good as the data in the peak list. It is important to understand what makes a good peak list and how the information is processed prior to a search.
This month's highlighted publication looks at the lost Giant Buddhas and the composition of the paint that once decorated them.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to create a summary report that combines the results from multiple searches.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Matrix Science Breakfast Workshops at ASMS
We are hosting two Breakfast Workshops at the ASMS meeting in Indianapolis and invite you to join us. Come hear about the latest applications and developments with database searching.
Both events are in the Indiana Convention Center, Room 137/138.
There is no charge for attending these meetings, but advance registration is required. Please go here for details and to register.
Monday 5 June, 7:00 am - 8:00 am
The Future of Peptide Identification by Spectrum Library Searching
presented by Stephen E. Stein, NIST
Integration of spectral library searching into Mascot Server
presented by Matrix Science
Tuesday 6 June, 7:00 am - 8:00 am
The Characterization of Therapeutic Proteins by Top-down and Bottom-up Approaches
presented by Paul W. Brown, Pfizer Worldwide Research and Development
New features in Mascot Server 2.6
presented by Matrix Science
|
 |
 |
 |
 |
|
Peak List Perfection
The peak list is the main currency of database search, and the information it contains is utilized in various ways. Some is critical and some is not used at all.
- Fragment charge is not required and, if present, is not used. Mascot always matches peaks to singly charged fragment ion masses. The instrument definition determines whether it also tries to match doubly charged. Usually, this depends on the precursor charge state.
- A default range of precursor charge states may be output by the peak picking software when it is unable to determine the precursor charge for a spectrum. These are treated collectively; the charge state for the highest scoring match is taken as correct and matches to other charge states are discarded.
- Short peptides will be ignored if they are shorter than the global setting MinPepLenInSearch. Peptide sequence matches shorter than MinPepLenInPepSummary are not sufficient evidence, by themselves, for the presence of a protein. At least one significant match to a longer peptide is required.
- Reliable de-isotoping can only be done on raw profile data. A good peak picking utility, such as Mascot Distiller, should be used to create a de-isotoped peak list. Mascot Server does not attempt to de-isotope the uploaded peak list.
Click here to read more about these issues and others in greater detail. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
GC/MS and proteomics to unravel the painting history of the lost Giant Buddhas of Bamiyan (Afghanistan)
Anna Lluveras-Tenorio, Roberto Vinciguerra, Eugenio Galano, Catharina Blaensdorf, Erwin Emmerling, Maria Perla Colombini, Leila Birolo, Ilaria Bonaduce
PLoS ONE 12(4): e0172990, April 5, 2017
The Giant Buddhas of the Bamiyan valley in Afghanistan dated from about 600AD and were the two largest clay statues in the world, until they were destroyed by the Taliban in 2001. They were fundamental pieces of art history not only because of their colossal dimensions, but also because of their influences in ancient Central Asia, being located in a valley that was connected to the Silk Road.
In this study the authors sought to understand the composition of the paint that once covered the statues by analyzing a number of clay fragments collected at the site. They fractionated the paint with three extractions and hydrolyses, giving saccharide, amino acid, and lipid/wax samples for GC/MS analysis. For protein analysis they used strong denaturing conditions followed by digestion and LC/MS/MS to identify the component proteins.
Lipids and waxes were not detected in any of the samples, while saccharides in a few, and proteinaceous matter in all the fragments. The proteomics analysis revealed that milk proteins were present and the authors were even able to conclude that cow and goat milk were the major paint binders.
|
 |
 |
 |
 |
|
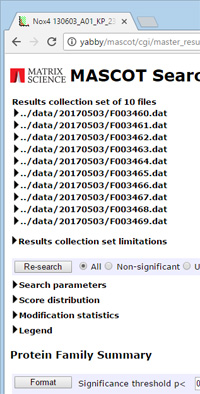
Mascot tip of the month
In Mascot 2.4 and later, it is possible to display a summary report that combines the results from multiple searches. This only makes sense if the searches use the same settings and the same database, such as a set of files analysed in a Mascot Daemon task. In fact, trying to mix searches with different settings will just give an error message when you try to view the report.
To create and populate a results collection, enter these URLs into the address bar of a web browser, remembering to replace 'your-server' with the name of your in-house Mascot Server
1. Create a results collection fileset:
http://your-server/mascot/cgi/client.pl?fileset_create
This returns the unique fileset_id to be used in the following URLs, e.g. fileset_2817400073158533
2. Add individual search results to the fileset:
http://your-server/mascot/cgi/client.pl?fileset_append; fileset_id=fileset_2817400073158533; file=../data/20150616/F001939.dat
If you wish to confirm which files are in the result set:
http://your-server/mascot/cgi/client.pl?fileset_list; fileset_id=fileset_2817400073158533
3. View the fileset report:
http://your-server/mascot/cgi/master_results_2.pl?fileset=fileset_2817400073158533
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|