|
To view this email as a web page, click here. |
 |
|
Welcome
We received a number of excellent submissions from our readers in response to our 'dark matter challenge', though most of the unidentified delta masses remain unexplained.
This month's highlighted publication describes a method for detecting blood-based biomarkers for amyloid-β, potentially showing a pathway to early Alzheimer's detection.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes how we can help you get up to speed faster with using Mascot Distiller to process your raw data.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Shedding light on dark matter
In a recent blog article, we asked our readers to help identify 20 unassigned mass differences found in open database searches. Thank you to all those who submitted comments and possible explanations.
The first successful identification was that 23.958 probably corresponded to an aluminium cation. Soon after, four other deltas were found to fit to combinations of amino acid residues, usually including K or R. With the benefit of hindsight, the explanation was obvious: the open database searches only allowed for one missed cleavage. If the matched peptide included two or more missed cleavages, this would be matched as a shorter peptide plus a modification. For example, K.SKLPKPVQDLIK.M would give a match as K.LPKPVQDLIK.M with an N-term mod of SK.
The remainder have proved more difficult to explain. In some cases, it is possible to find an elemental composition within experimental error, but not a compelling chemistry. For example, 306.0952 might be glycation: addition of 2 Hexose and loss of water. The difficulty with this is the absence or near absence of similar modifications, such as addition of 1 hexose with loss of water or addition of 2 Hexose without loss of water. These are abundant modifications found on a variety of peptide sequences. If they were combinations of modifications, we would expect to see the individual modifications at even higher abundance.
This competition has certainly demonstrated that figuring out the chemistry for an unknown delta is very difficult. The unexplained positive deltas greater than 5 Da that were not in Unimod have been added to Unimod with the prefix Unknown, e.g. Unknown:302. If you perform an error tolerant search and these deltas are present, strong matches will help narrow down the site specificity. Please share such results by adding your feedback as comments to either the original blog article or the results round-up.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
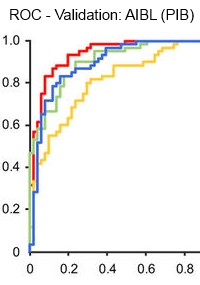
High performance plasma amyloid-β biomarkers for Alzheimer's disease
Akinori Nakamura, Naoki Kaneko, Victor L. Villemagne, Takashi Kato, James Doecke, Vincent Dore, Chris Fowler, Qiao-Xin Li, Ralph Martins, Christopher Rowe, Taisuke Tomita, Katsumi Matsuzaki, Kenji Ishii, Kazunari Ishii, Yutaka Arahata, Shinichi Iwamoto, Kengo Ito, Koichi Tanaka, Colin L. Masters & Katsuhiko Yanagisawa
Nature, Published online: 31 January 2018
In order to move toward a readily available test for early stages of Alzheimer's disease, the authors have identified and validated blood-based biomarkers for amyloid-β proteins. Using immunoprecipitation with MALDI-TOF MS, they were able to establish that the biomarkers in the IP-MS assay were potentially clinically useful candidates as surrogates for brain amyloid-β burden.
Their retrospective cross-sectional study tested these markers in a discovery data set from the Japanese National Center for Geriatrics and Gerontology (121 samples), and was externally validated using an independent data set derived from the Australian Imaging, Biomarker and Lifestyle Study of Ageing (252 samples).
They also looked at the relationships between the plasma biomarkers and two accepted approaches for amyloid-β determination, positron emission tomography and cerebral spinal fluid biomarkers. These results demonstrate that the three different types of Aβ related biomarkers (plasma and CSF Aβ, and PET imaging), are highly correlated with each other, indicating that plasma Aβ biomarkers are strongly linked with the Aβ status of the CNS.
|
 |
 |
 |
 |
|
Mascot tip of the month
Learning to use a new software package of any complexity is always a bit of a struggle, and Mascot Distiller is no exception. Particularly when the data are being quantified; everything has to be just right to get good results: the initial peak picking, the search conditions, and the settings for the final quantitation step.
We try our best to provide tutorial-style help, but it can never be good enough. The sheer variation in raw data and processing pathways makes it impossible to have useful defaults for every setting. High resolution or low? Saved as profile or centroids? Quantitation off MS1 or MS2 or MS3? SILAC or 18O or 13C metabolic or label-free or iTRAQ or TMT or dimethylation or ...
The learning curve is much less of a challenge if you have a good model on hand, where reasonable choices have been made for these myriad settings. You can immediately see how things are supposed to work and focus on optimisation.
We are happy to offer such assistance. Contact support with some basic information about the experiment, e.g. organism, protease, type of labelling, etc. We will send an upload link to our dropbox. Send us one file, or two if it is label free. (No more - we don't need a huge data set to illustrate suitable settings.) We will process your file and send back a Distiller project file, which gives you a good starting point for processing the rest of your data.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|