|
To view this email as a web page, click here. |
 |
|
Welcome
We mark 25 years since the first publications describing Peptide Mass Fingerprinting, one of the key methods of MS-based proteomics.
This month's highlighted publication presents a new approach to profiling of glycopeptides using database search.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month lists useful sources of information about specifying hardware for Mascot Server and Mascot Distiller.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
A quarter-century of MS-based protein ID
Protein identification by searching mass spectral data against a protein sequence database is now such a common tool that it is hard to imagine not having it. In fact, this year marks the 25th anniversary of the first publications.
Five groups laid the foundations for today's proteomics research, describing the method of peptide mass fingerprinting. This was soon extended to the use of MS/MS for peptide identification.
The field has come a long way since SwissProt contained only 30k entries and no genomes had yet been sequenced. Please go here to read more about the early days and the researchers that started it all.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
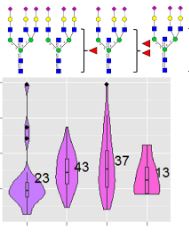
Large-scale intact glycopeptide identification by Mascot database search
Ravi Chand Bollineni, Christian Jeffrey Koehler, Randi Elin Gislefoss, Jan Haug Anonsen & Bernd Thiede
Scientific Reports, volume 8, Article number: 2117 (2018)
The authors have developed a novel method for identifying intact glycopeptides in a high-throughput and batch-wise manner. They created a custom peptide database in which known glycan structures were linearized and then encoded using the one letter codes O (GlcNAc, GalNAc), J (Galactose, Mannose) and U (Neu5Ac). Additional monosaccharides could be encoded as variable modifications to these codes. By assigning the appropriate mass values and attaching the glycan sequence at the peptide N-terminus, Mascot was enabled to match glycosidic cleavage ions from both N-linked and O-linked glycans as pseudo y-ions.
Serum samples from controls and patients diagnosed with prostate cancer were analyzed to validate the approach. Samples were digested with trypsin and desalted with ZIC-HILIC SPE, enriched with TiO2 beads, followed by LC-MS analysis. The intact glycopeptide mass spectra were submitted to the Mascot search engine for identification and relative quantification with Mascot Distiller. The data was searched against a custom glycoprotein database.
A total of 257 glycoproteins were identified from the 24 serum samples. Within these 257 glycoproteins, a total of 970 unique glycosylation sites and 3447 non-redundant glycopeptide variants were identified.
|
 |
 |
 |
 |
|
Mascot tip of the month
PC hardware specification has been mentioned in past newsletters, but it's a topic that comes up frequently, so here is a summary the main sources of information.
The hardware requirements for Mascot Server and Mascot Distiller are very similar. Both are processor bound for much of the time. The main difference is that a Mascot Server licence limits the number of cores used for searching. A 1 cpu licence will use a maximum of 4 physical cores or 8 logical cores (if the cores are hyperthreaded) for searches. It's useful to have some additional cores for reports, utilities, and the operating system, but you won't see much difference in search speed by adding many more cores over and above those covered by the licence.
Mascot Distiller, on the other hand, is free to use all the cores it can find. The number used for an individual raw file is capped to avoid file access becoming a bottleneck, but when processing files in multiple Daemon tasks or as a multi-file project, you may see all cores saturated. This means that you can get competition for processor time if you install Mascot Server and Mascot Distiller on the same PC.
These articles and help pages are the most relevant to hardware specification:
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|