|
To view this email as a web page, click here. |
 |
|
Welcome
To celebrate our 20th anniversary, we have a special offer: 20% off all Mascot Server updates.
We are hosting two ASMS breakfast workshops on June 4th and 5th in San Diego and invite you to join us.
We are also pleased to announce the release of Mascot Distiller 2.7, which is now much faster at processing most profile data. We describe a Distiller workflow for MS3 reporter ion quantitation.
This month's highlighted publication shows a simplified method for identifying protease substrates via their N-termini.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
20-20 Special offer
This year marks the 20th anniversary of Matrix Science. To celebrate, we are offering 20% off all Mascot Server updates. This limited-time offer is for orders received by 31st July 2018.
Many essential new features have been added in recent releases, such as Percolator support in 2.3, Database Manager in 2.4, 64-bit Mascot Daemon in 2.5, and integrated spectral library searching in 2.6.
If you have a Mascot Server licence that has fallen behind the current release, 2.6, just e-mail sales@matrixscience.com requesting a quotation and we will automatically apply this discount. More information here.
|
 |
 |
 |
 |
|
Matrix Science Breakfast Workshops at ASMS
Please join us at our annual User Meetings at the upcoming ASMS Conference in San Diego, California. These breakfast workshops take place on June 4th and 5th at the San Diego Convention Center, where we will describe the latest tools and techniques for database searching. We also have two featured speakers presenting their exciting applications.
Monday 4 June, 7:00 am - 8:00 am
Expanding the landscape of human phosphorylation
presented by Claire Eyers, Centre for Proteome Research, U. Liverpool
Mascot Distiller gets up to speed
presented by Matrix Science
Tuesday 5 June, 7:00 am - 8:00 am
Identification of Iodine-containing Alkylation Reagent related Artifacts and Polymer Contaminations in Proteomics Samples
presented by Dominic Winter, Institute for Biochemistry and Molecular Biology, U. Bonn
Using spectral libraries to pick a peck of pickled peptides
presented by Matrix Science
Please go here to register and reserve your spot as seating is limited. Breakfast will be served. |
 |
 |
 |
 |
|
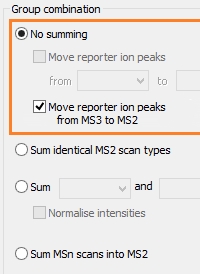
MS3 reporter ion quantitation with Mascot Distiller
When using reporter ion quantitation methods, such as iTRAQ or TMT, interfering species in the reporter ion region can skew the results. Many researchers resort to further fragmenting a major peak in the MS/MS spectrum and using the reporter ions in the MS3 spectrum, which are then mostly free of interference.
Mascot Distiller supports this workflow. The MS3 reporter ions are spliced into the MS/MS spectrum, replacing the MS2 reporter ions. This is better than simply summing pairs of MS3 and MS2 spectra.
We demonstrated this with a published TMT 6-plex dataset. Using only the MS3 reporter ions gave a peptide ratio of 2.99 (against expected value of 3.0) while summing the entire spectra gave a value of 2.70. Quantifying the MS2 data alone gave 1.99.
Please go here to read more.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Simple, scalable, and ultrasensitive tip-based identification of protease substrates
Gerta Shema, Minh T. N. Nguyen, Fiorella A. Solari, Stefan Loroch, A. Saskia Venne, Laxmikanth Kollipara, Albert Sickmann, Steven H. L. Verhelst and Rene P. Zahedi
Molecular & Cellular Proteomics, (2018) 17 826-834
The authors have developed a new method for the identification of novel N-termini formed by the action of proteases upon various protein substrates, revealing information about the regulatory proteolytic networks in health and disease. The method relies on the enrichment of the N-terminal peptides followed by MS identification and quantification.
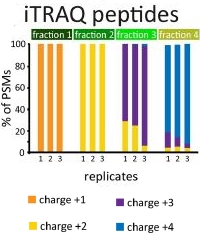
The method, charge-based fractional diagonal chromatography in a pipette tip or ChaFRAtip, utilizes well-established protein chemistry and minimal equipment: A pipette tip with a cellulose frit, strong cation exchange chromatography beads and common buffers. In conjunction with iTRAQ-8-plex labeling, the method can quantify over 2,000 N-terminal peptides across eight samples using around 4 µg of protein per condition/channel.
The authors compared the original LC/MS methodology to ChaFRAtip using SH-SY5Y cells treated with staurosporine to induce apoptosis. They achieved a similar level of technical reproducibility for both methods and quantified 975 ±24 (HPLC) and 783 ±71 (tips) unique N-terminal peptides at 1% false discovery rate.
|
 |
 |
 |
 |
|
Mascot Distiller 2.7
We are pleased to announce the release of Mascot Distiller 2.7. The most important change is that it is no longer necessary to 're-grid' spectra if the mass values of the data points do not represent a continuous, linear mass scale. This makes peak picking very much faster for compressed profile data, where baseline data points have been discarded, and avoids the possibility of distorting peaks by specifying a mass scale that is too coarse.
You can download Mascot Distiller from a link on the support page. If you are using an earlier version, remember that Mascot Distiller updates are free! If you wish to evaluate Mascot Distiller, send an email request to support@matrixscience.com for a product key that can be registered on-line to obtain a 30 day licence. Without registration, or on expiry of the evaluation, Mascot Distiller operates in viewer mode.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|