|
To view this email as a web page, click here. |
 |
|
Welcome
We are looking forward to seeing many of you at the upcoming World HUPO meeting in Florida, where we are offering free advice sessions.
Below, we illustrate the flexibility of Mascot Distiller by showing how it can be customised for a novel method for quantifying cysteine oxidation.
This month's highlighted publication concerns a method for identifying protein targets of catechol estrogens.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes how to fix broken search result links in Mascot Daemon.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Free consultations at World HUPO
- Have a question about error tolerant searches or post-translational modifications?
- Thinking of trying a new quantitation method?
- Want to add a new database to Mascot Server or set up Mascot security?
- Need advice on how to interpret your search results?
We are offering free advice on the best use of Mascot Server, Mascot Daemon and Mascot Distiller at the 17th World HUPO Conference in Orlando, Florida (30 September to 3 October).
Thirty minute time slots are available for a maximum of two people at a time. There won't be time to run long searches or process large raw files so, once you have booked a slot, you are welcome to send us data in advance. If you wish to do this, contact support@matrixscience.com to discuss what files to send.
Location will be the Matrix Science exhibition booth, which is #307. To book a slot, follow this link.
|
 |
 |
 |
 |
|
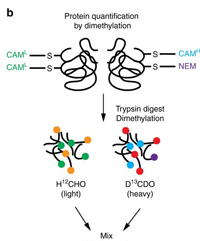
Quantitation of cysteine oxidation
Earlier this year, a group from the University of Glasgow described a novel quantitation method based on light and heavy iodoacetamide labels, which they named SICyLIA. At its core, SICyLIA is a precursor protocol with two components. However, there’s a twist: reversibly oxidised cysteine thiols are reduced and blocked with N-ethylmaleimide (NEM) after pooling. Light and heavy peptides with more than one cysteine may well have NEM-blocked cysteines, and there may be peptides carrying only NEM.
In the published study, peptides with both a label and NEM-blocked cysteine (3-4%) were omitted from quantitation due to software limitations. But it's easy to define a quantitation method for Mascot Distiller that accounts for NEM-blocking.
In most Precursor quantitation methods, the light and heavy components are modelled as exclusive modifications, which is a choice of fixed modifications. This means all potential sites for labelling must be identically modified.
For SICyLIA, we want to allow zero or more NEM-blocked cysteines. Easily done: define the components using variable modification groups, which allows Mascot to match peptides in all eight cases (unmodified; NEM; light; heavy; light + NEM; heavy + NEM; light + heavy; light + heavy + NEM). Distiller assigns a peptide to the light component if it carries at least one light CAM, and to the heavy component if it carries at least one heavy CAM. If the peptide has both light and heavy CAM, it correctly fits neither component.
This provides an excellent illustration of the flexibility of Mascot Distiller quantitation. A more complete description
can be found here.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
In Situ Click Reaction Coupled with Quantitative Proteomics for Identifying Protein Targets of Catechol Estrogens
Huei-Chen Liang, Yi-Chen Liu, Hsin Chen, Ming-Chun Ku, Quynh-Trang Do, Chih-Yen Wang, Shun-Fen Tzeng, and Shu-Hui Chen
J. Proteome Res. Publication online: June 13, 2018
Catechol estrogens (CEs), major metabolites of estrogens, are an important set of electrophiles that form stable covalent conjugates with endogenous proteins. These covalent modifications may induce cell signaling responses and influence the metabolic or gene regulation. In this study, the authors developed a click reaction/MS-based strategy to identify cellular protein targets of CEs.
Liver microsome samples were treated with ethinylestradiol as the precursor probe, and the CE metabolites were produced in situ by the enzymes present in the microsomes. After 24 hr. the CEs-protein complexes were affinity purified using click chemistry with a biotin azide linker. The samples were digested by trypsin, stable isotope labeled via reductive amination, and analyzed by LC/MS.
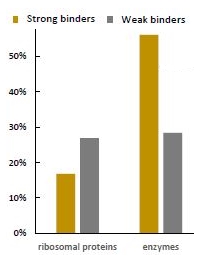
A total of 334 liver proteins were identified and 274 proteins were classified as strong binders. Many of the identified strong binders are involved in cellular redox processes or detoxification activities.
|
 |
 |
 |
 |
|
Mascot Tip
Moving Mascot Server to new hardware is not difficult, and there are migration procedures that provide step by step instructions. Ideally, you would also transfer the host name from the old server to the new one, so as to preserve bookmarks and references to the server in other software. But, this can be complicated, and may not be an option in your organisation
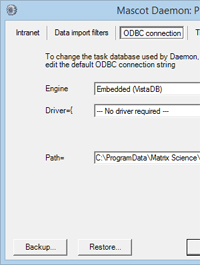
More seriously, when the server URL changes, all of the links to search results in Mascot Daemon will break. This can be annoying if Mascot Daemon is your main user interface. Provided this is Mascot Daemon 2.6, there is an easy fix:
- In Mascot Daemon, choose Preferences from the Edit menu
- On the ODBC connection tab, select Backup
- The task database will be exported to an XML file
- Open the XML file in a text editor (not a word processor)
- Use search and replace to change all instances of the old server host name. For example, change http://lab_server/mascot/ to http://new_server/mascot/
- Save the changes, then import the file back into Mascot Daemon by choosing Restore
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|