|
To view this email as a web page, click here. |
 |
|
Welcome
We explain when and why you might wish to switch your Mascot Server into cluster mode.
This month's highlighted publication shows how to obtain a 2-fold increase in phosphopeptide ID by reducing ion suppression.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes how to re-build the Mascot Server search log.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Should you switch to cluster mode?
If you have a Mascot Server license for several CPUs, there comes a point where it is more efficient and cost-effective to use a cluster of 'commodity' PCs, because processors with very large numbers of cores can be expensive and may require exotic system board hardware. Cluster mode is a standard feature of Mascot Server; you don't need a special version.
- For licenses of 3 or 4 CPU, you could choose a single machine, possibly with dual processors, to keep things simple, although you might achieve the same performance at lower total cost with a couple of single processor PCs.
- For licenses of 5 CPU or more, a cluster is often the most practical option. All machines in a cluster should have processors of the same speed. Otherwise, the PC with the slower processor(s) will become a bottleneck.
- For a large cluster, it may be best not to run searches on the master (head) node of the cluster, but leave it free to run the web server, handle database updates, and generate reports. This ensures the server will be responsive even when there are several searches running.
- Since a Mascot licence is for a fixed number of cores, it is the single thread benchmark for the processors that matters most.
Go here to read more about hardware considerations for maximizing performance while minimizing cost.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
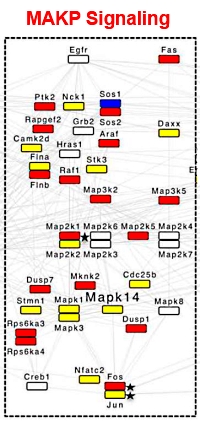
Global ion suppression limits the potential of mass spectrometry based phosphoproteomics
Roland Felix Dreier, Erik Ahrne, Petr Broz, and Alexander Schmidt
J. Proteome Res., Article ASAP, published online November 2, 2018
The authors evaluated the impact of phosphorylation on peptide identification by LC-MS. They used an efficient dephosphorylation protocol to directly compare identified phosphopeptides with their unmodified counterparts. The samples in this study were generated in large-scale phosphoproteomics studies including frequent missed cleavages, large abundance differences and common contaminations, as opposed to the standard phosphopeptide mixtures sometimes employed.
A mix of two potent phosphatases (lambda- and alkalinephosphatase) was applied to TiO2-enriched samples from whole HeLa S3 cell lysates. The phosphatase mix removed virtually all phosphorylations after only 1 hour of treatment (>99.7% complete). When looking at the LC-MS chromatograms, the authors noticed an approximate 10-fold increase in precursor ion intensities after phosphatase treatment. Using the standard phosphoproteomics approach, they quantified 6,607 phosphopeptides from the enriched samples. When implementing the additional phosphatase treatment, precursor ion intensities were strongly enhanced and the number of quantified peptides increased by a factor of 2.3 to 15,494 peptides.
The immune signaling pathways engaged during Salmonella infection of macrophages were investigated and it was determined that the use of this phosphatase treatment doubled the number of proteins in the protein-protein interaction analysis. Sample matrix related ion suppression is the main driver of the reduced MS response/identification rates observed. This led to MS sequencing of additional peptide precursor ions, which were almost completely suppressed in the original phosphopeptide enriched sample.
|
 |
 |
 |
 |
|
Mascot Tip
The Mascot Server search log is the main way to locate the result report for an existing search. There are times when you might need to rebuild the search log:
- The log file is lost or damaged
- You delete old result files to save space and don't want to see them listed in the log
- You merge the result files from two Mascot Servers into a single, new system and want all the search history in one place
Mascot includes a utility - ms-makesearchlog.exe in the Mascot bin directory - that scans the result files on the server to create a new log file. Ideally, this is done at a quiet time, such as a weekend, to avoid missing new result files that are created while the utility is running.
ms-makesearchlog.exe is a command line utility with no arguments. It ignores the paths defined in mascot.dat and is hard-coded to scan for result files under ../data/ and create a searches.log file in ../logs/. The existing log file is renamed to searches.bak.
If the program files are in one location but the result files in another, you may need to create logs and bin directories at the same level as the data directory and copy the executable to the bin directory. Once processing is complete, move the new log file to the active logs directory.
The other 'gotcha' to be aware of is that searches.log needs to be writeable by Mascot Monitor and readable by CGI applications. Either compare the permissions / security settings on the old and new files or just make the new file world writeable (chmod 666 in Linux or full control for the local SYSTEM group in Windows).
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|