|
To view this email as a web page, click here. |
 |
|
Welcome
A recent study illustrates the challenges of benchmarking protein inference.
This month's highlighted publication describes how a customized database was created to investigate photosynthesis in Norway spruce.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month may come in useful if your server is getting overloaded with long searches.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Benchmarking protein inference
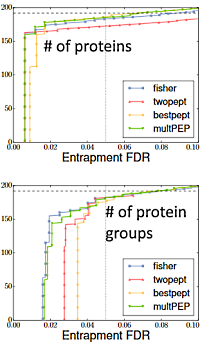
A recent blog article looked at an ABRF research group study aimed at benchmarking different approaches to protein inference. The samples for this study were prepared from protein epitope signature tags expressed in E. coli by selecting 191 overlapping pairs so that each pair would have some unique and some shared tryptic peptides. The pairs were divided into two pools, A and B, and a third pool was created by combining A and B.
Samples from the pools were spiked into an E. coli cell lysate for analysis. Participants were also supplied with Fasta files containing the sequences in the two pools plus a collection of decoy sequences that could be used to estimate protein FDR.
While the sample is more realistic than many, the database is artificial, in that it contains perfect representations of all the target sequences. This is not the case for most real-life searches, which use a public database in which the sequence of a particular protein may differ from that in the sample.
This has particular relevance to the choice of whether shared peptide matches should be included or excluded from protein inference. A realistic, non-identical database might contain two homologous protein sequences, maybe 90% identical. Unless the coverage is almost complete, excluding shared matches will mean that neither protein appears in the result report, which cannot be a good thing.
Go here for more details of how the database influences the search results and how the Mascot protein family summary report handles ambiguity.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
The unique photosynthetic apparatus of Pinaceae: analysis of photosynthetic complexes in Picea abies
Steffen Grebe, Andrea Trotta, Azfar A Bajwa, Marjaana Suorsa, Peter J Gollan, Stefan Jansson, Mikko Tikkanen, and Eva-Mari Aro
J. Exp. Botany, 70 3211-3225 (2019)
The authors are investigating the survival strategies of evergreen species in the harsh environmental conditions of boreal forests. To this end, they have elucidated the protein composition of the photosynthetic light-harvesting and energy conversion machinery of the Norway spruce, Picea abies.
They performed the analysis using 2D SDS-PAGE and LC-MS/MS together with a custom database which was constructed from chloroplast- and nuclear-encoded protein sequences which were merged with protein sequences derived from transcripts. These transcript nucleotide sequences were selected by tBLASTn searches against known thylakoid associated protein sequences from reference species. The candidate sequences were translated to protein sequences, and both contaminant and truncated sequences were removed. This led to the annotated spruce thylakoid protein database which was combined with the chloroplast- and nuclear-encoded protein sequences to form a merged protein database.
To compare the composition of the light-harvesting complexes (LHC) of Norway spruce with other land plants, they classified all available LHC homologues from genomes and transcriptomes of 84 plant species, utilizing homology between selected diagnostic sequence regions. This analysis identified major differences in LHC composition between members of Pinaceae and other plants.
|
 |
 |
 |
 |
|
Mascot Tip
Is your Mascot Server struggling to keep up with its workload? Obviously, we'd be delighted if you bought a licence upgrade, to use more processors. On the other hand, you may be able to alleviate the situation for free by tweaking priorities.
If you go to the Database status page, and click on a database, you get a list of current and recent searches against that particular database. Click on a search that is still running, and you'll find controls to change the priority of the search, pause it, or even kill it. If there are long searches that are not urgent, such as other people's searches, they can be given a low priority, so that they only get significant processor time when the system would otherwise be idle. The lower the value, the lower the priority.
If Mascot security is enabled, the default priority, which is also the highest priority, can be set differently for different groups. Users can still decrease the priority for their non-urgent jobs, but they cannot increase it above their limit. Mascot security also allows you to set limits at group level on maximum search time, the number of concurrent searches per user, and all of the factors that influence how long searches might take (except precursor tolerance).
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|