|
To view this email as a web page, click here. |
 |
|
Welcome
Diethylation labelling for quantitation has all the advantages of dimethylation without the deuterium effect.
This month's highlighted publication demonstrates a new method for analyzing previously intractable protein aggregates.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month highlights where to find help for migrating your Mascot Server to a newer version of Windows.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Diethylation, dimethylation and the deuterium effect
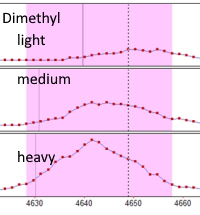
The 3-plex dimethylation method, which involves the labeling of N-terminus and Lysines with light, medium and heavy dimethylation reagent, is complicated by the 'deuterium effect'. This is where the different numbers of deuteriums between the light, medium and heavy labels result in shifted retention times for the peptides. Mascot Distiller can accommodate this by allowing for the elution time to shift within a defined window, so as to align the peaks for measurement. The down side is an increase in computation time and the possibility of misalignment, which becomes most likely when one or more XIC peaks are weak or noisy.
Diethylation is a new, 3-plex label that avoids the deuterium effect on retention time. This new method will need to be added to your Mascot Server, but this is easily done. The 'light' diethyl modification is present in Unimod, but the medium and heavy variants are not, so you'll need to add the medium and heavy component modifications to your local copy of Unimod. You can then define the Diethylation quantitation method using the Mascot Server configuration editor.
A comparison of these methods, using data from PRIDE, show that diethylation compares very favorably with dimethylation.
Both methods gave median ratio values very close to the expected ratios, but more peptides could be quantified using diethylation, giving better precision on the measurements
Go here to read the details of the study as well as how to implement diethylation on your Mascot Server.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
An Ionic Liquid-Based Sample Preparation Method for Next-Stage Aggregate Proteomic Analysis
Masato Taoka, Koji Horita, Takahiro Takekiyo, Takamasa Uekita, Yukihiro Yoshimura, and Tohru Ichimura
Analytical Chemistry 91 13494-13500 (2019)
This paper describes a new method to analyze aggregated proteins using ionic liquids for solubilization. Prior work in this area has often suffered from large sample requirements as well as complex, labor-intensive protocols.
Using heated egg whites as a test case, they dissolved the aggregated proteins with an optimized mixture of 1-butyl-3-methylimidazolium thiocyanate/NaOH. This provided complete dissolution of egg white proteins within 10 minutes. They performed further sample cleanup prior to digestion by concentrating the proteins onto reversed-phase support and removing the contaminants with buffer washing. LC-MS/MS analysis showed this method to produce 79 peptides and 8 proteins in comparison to standard approaches such as SDS, urea, formic acid which yields 10-20 peptides and 1-3 proteins
The authors applied their technique to the model nematode C. elegans, which produces protein aggregation upon aging. In comparison to a formic acid solubilization, this ionic liquid method identified 176 vs 26 proteins, including many more transmembrane proteins.
|
 |
 |
 |
 |
|
Mascot Tip
14 January 2020, the end-of-life date for Windows 7 and Server 2008, is fast approaching. This has prompted many people with Windows-based Mascot Servers to migrate their systems to Windows 10. Note that a detailed migration procedure is available.
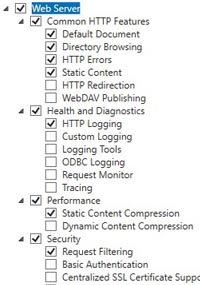
One of the critical installation steps is to enable and configure IIS, Microsoft's web server. If you are running an old version of Mascot, you won't find screen shots for the latest versions of Windows in the manual. To remedy this, we have added a page of IIS screen shots to the the support material on our web site.
It is essential that IIS is configured correctly before Perl or Mascot is installed. Trying to correct the configuration after Mascot has been installed can be very difficult.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|