|
To view this email as a web page, click here. |
 |
|
Welcome
We want to make the public Mascot Server as useful as possible to those of you having to work remotely during the current emergency, so we have added new databases and increased the maximum search size. Go Here to read more.
Protein FDR can be controlled by adjusting peptide FDR or filtering out one-hit wonders - which works best?
This month's highlighted publication describes a new enzyme to produce peptides with a basic residue at the N-terminus.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month gives further details about the recent changes to the public Mascot Server.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Protein FDR in Mascot Server 2.7
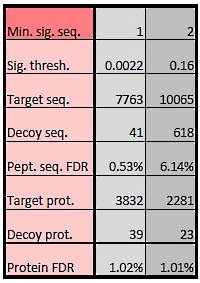
One of the new features in Mascot Server 2.7, now running on our web site, is an estimate of protein FDR. The basis is the number of proteins inferred in the target database compared with the number in the decoy database. Though this is similar to peptide FDR, there are several other factors that must be considered.
- Only peptide sequence matches (PSMs) with significant scores are used as evidence for proteins, and proteins with shared PSMs are grouped into families.
- A family member may represent multiple same-set proteins, one of which is given prominence as the anchor protein.
- The protein count used for FDR is a count of family members. That is, if the report contains 2 families, one with 4 members and the other with a single member, this counts as a total of 5 proteins.
- A protein identification is considered to be a true positive if it contains at least one true positive PSM. A protein is a false positive only when all of its PSMs are false positives.
One way to control protein FDR is by adjusting peptide FDR. There is a second way to control protein FDR: changing the minimum number of significant unique sequences. At the default setting of 1, we are reporting 'one-hit wonders'. For large searches, conventional wisdom is that it is safer to exclude these, as explained in an earlier article. Yet, in many cases, we can report more proteins for a given protein FDR by including one-hit wonders. The table on the right shows a typical example.
To read about these considerations in greater detail, please go here.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Tryp-N: A Thermostable Protease for the Production of N-terminal Argininyl and Lysinyl Peptides
John P. Wilson, Jonathan J. Ipsaro, Samantha N. Del Giudice, Nikita Saha Turna, Carla M. Gauss, Katharine H. Dusenbury, Krisann Marquart, Keith D. Rivera, and Darryl J. Pappin
J. Proteome Res. Publication Date: March 6, 2020
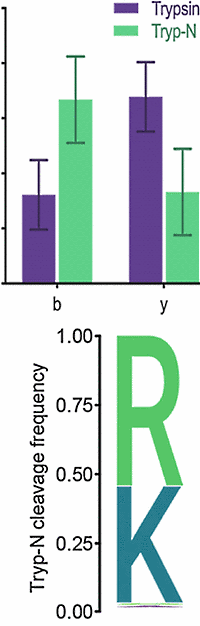
The authors undertook an investigation to identify enzymes that produce peptides with basic centers exclusively on their N-termini, producing mainly b-ions and simplifying peptide fragmentation and assignment. They computationally screened enzymes to identify proteases and putative proteases, which might possess the desired N-terminal cleavage specificity.
Candidate genes were synthesized, processed via in vitro transcription and translation, with those that displayed significant digestion characterized for protease specificity by MS of the resultant peptides. They further characterized candidates for protease activity and specificity, pH sensitivity, detergent tolerance, and MRM sensitivity.
Tryp-N was found to show high activity over a broad pH range and buffer conditions, and had a ≥95% specificity for arginine and lysine. Because charge is mostly retained by the N-terminal fragments, the Tryp-N generated peptides often exhibit lower limits of detection during MRM analyses as compared to their tryptic peptide counterparts.
|
 |
 |
 |
 |
|
Mascot Tip
As explained in this news item, the NCBI non-identical protein database, NCBIprot, is now only available on request. The reason is its sheer size: 227 million sequences and 82 billion residues, making it 400 times larger than SwissProt. Even with a taxonomy filter, the huge number of entries will severely limit sensitivity. For example, there are 12 million sequences in Fungi and 85 million in Proteobacteria.
If you are studying an organism that is poorly represented in SwissProt and for which there is no Uniprot proteome with decent coverage, you have no choice, and we are happy to provide a login to run searches of NCBIprot. If necessary, we will add a category to the taxonomy filter, so that a suitable genus or family can be selected.
If you are studying an organism for which there is a Uniprot proteome with good coverage, this is the ideal search target. The search will be fast and the result report clear to interpret. These proteomes are all configured to include variants, and if the results are good, you can even run an error tolerant search to pick up additional single residue substitutions. If we don't have a proteome database for your organism of interest, and Uniprot has a proteome with at least 75% coverage, contact us with a request that it be added.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|