|
To view this email as a web page, click here. |
 |
|
Welcome
Welcome to Mascot Newsletter, our new monthly communication aimed at providing helpful information on protein identification, characterization, and quantitation. This month we have a few ideas on improving search speed, which is always a concern.
In addition, each month we will feature a journal article that uses Mascot Server or Mascot Distiller in a novel way or for an important or challenging application. If you have a recent publication that you would like us to consider for an upcoming Newsletter, please send us a PDF or a URL.
Finally, we would like to remind you that Mascot version 2.5 was released in July, and we have a brief overview of the important new features and how you can update.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
How long should my search take?
"I wish my searches were faster..." is a common refrain from many researchers, wondering which factors are most responsible for the logjam. We have a few suggestions here to help you sort through some of these variables and potentially gain that extra speed you were hoping for:
1. The effect of database size is approximately linear with search time. Remember that comprehensive databases grow larger with every update.
2. Search time is proportional to the peptide mass tolerance. That said, fragment ion tolerance has little or no effect on search time.
3. A no-enzyme search may take 100 times as long as a search with tryptic specificity. Using an error tolerant search is a much faster and more sensitive way of matching semi-specific and non-specific peptides.
4. Don't set the allowed number of missed cleavages to an unnecessarily high value. Changing the setting from zero to one missed cleavage approximately doubles the search space.
5. Fixed modifications have no effect on search times, but variable modifications cause a geometric increase in search times. Only modifications that are very abundant should be specified as variable modifications.
6. Quantitation protocols that are handled by the search engine, such as iTRAQ and TMT, add to the time taken to display the report. Best to set normalization to none in the method. Once the report is loaded, you can check whether everything looks OK and the various format options are set correctly before finally setting normalization as required and going for a well-earned cup of coffee.
You can click here to see more ideas and details on optimizing your search speed. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Tracking Cancer Drugs in Living Cells by Thermal Profiling of the Proteome
Mikhail M. Savitski, Friedrich B. M. Reinhard, Holger Franken, Thilo Werner, Maria Fälth Savitski, Dirk Eberhard, Daniel Martinez Molina, Rozbeh Jafari, Rebecca Bakszt Dovega, Susan Klaeger, Bernhard Kuster, Pär Nordlund, Marcus Bantscheff, Gerard Drewes.
Science 3 October 2014, Vol. 346, no. 6205
This paper demonstrates a new approach for the unbiased measurement of drug-target binding, illustrating the interaction of anticancer drugs with human leukemia cells. By measuring the changes in the thermal profiles or "melting points" for over 7,000 proteins, the authors were able to identify over 50 targets for the kinase inhibitor, staurosporine. Additionally, the heme biosynthesis enzyme ferrochelatase was inhibited, suggesting this as a cause for the phototoxicity observed with vemurafenib and alectinib. It is expected that thermal proteome profiling can be extended to membrane proteins as well as tissues from animal studies or clinical biopsies. In a press release on the study, Bernhard Kuster of Technische Universität München and the German Cancer Research Center said, "We hope to use this method in the future to explain or even predict many adverse effects." |
 |
 |
 |
 |
|
Mascot Server 2.5 is now available
Version 2.5 of Mascot Server was released in July, and there are new features that you may find very helpful in your work. If you were under warranty or support in July, you were emailed with download information for your free update. If this message was missed for some reason, please contact us to obtain your update.
Remember to check the support page for known issues and documentation updates. If you plan to move to new hardware at the same time as updating to Mascot 2.5, consult our step-by-step migration procedures.
Important new features include:
Chimeric spectra
With high resolution data, it is possible for two overlapping isotope distributions to be cleanly resolved. If two precursors return different matches, both are reported. If two precursors return the same match, this is only reported once, using the closest m/z value.
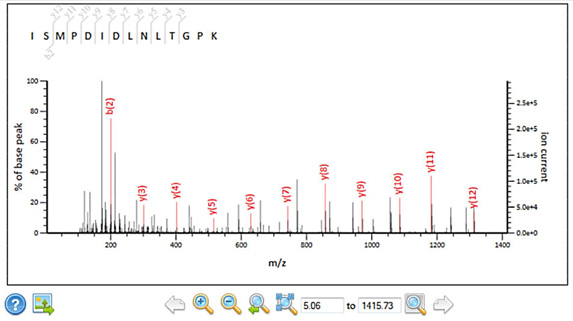
New spectrum viewer
Peptide view has been upgraded with an interactive, SVG spectrum viewer that supports zoom and pan. |
 |
 |
|
Additional improvements
New modifications and edits to existing modifications are now stored separately, and you can make modifications private to selected user groups.
Mascot Daemon has been totally re-written and is now available in both 64-bit and 32-bit versions.
Counts of modifications are displayed at the top of the Protein Family Summary, making it easier to spot interesting modifications or assess which modifications are the most abundant.
A Fasta file can be uploaded through Database Manager, allowing you to add a new sequence database when you only have a web browser (HTTP) connection to a Mascot Server.
More information in this presentation |
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|