|
To view this email as a web page, click here. |
 |
|
Welcome
We are hosting our annual
Mascot Workshop and User Meeting
at the ASMS meeting in St. Louis on May 31. Please read the details below and register to reserve your place.
Many people are looking to virtual machines as way to increase efficiency. We present some pointers on configuring a VM for Mascot.
This month’s highlighted publication describes a method to measure phosphorylation occupancy across complex protein samples.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns migrating to new hardware.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Mascot Workshop and User Meeting 2015
Please join us at our Annual Mascot Workshop and User Meeting at the upcoming ASMS meeting in St. Louis, May 31. Learn of the latest approaches and applications for database searching, identification, and quantification of proteins and peptides.
The meeting will take place at the Crowne Plaza St. Louis – Downtown from 8:30am to 12:30pm. There is no charge for attending this workshop, but advance registration is required. Seating is limited, so you should register as soon as possible.
Go Here for details and registration. |
 |
 |
 |
 |
|
Running Mascot in a Virtual Machine
Virtualization is an effective way to reduce IT expenses while boosting efficiency and agility. It can offer a host of advantages such as consolidation, elastic provisioning, high availability / disaster recovery, multi-tenant isolation, and legacy OS support.
Some of these benefits could apply to a production server running Mascot searches 24/7, but installing Mascot Server in a VM requires careful consideration to resource allocation. Factors such as:
- A Mascot license is priced according to the number of processor cores available for searches so it is important to choose fast hardware.
- You need to ensure that the virtual processors are correctly configured.
- Search speed depends heavily on the sequence database files being held in memory during the search, so it’s important that the VM is given access to as much physical RAM as practical.
- Storage arrangements can also impact Mascot performance, so care is needed to avoid significant bottlenecks.
You can read more details here on how to optimize your VM setup. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
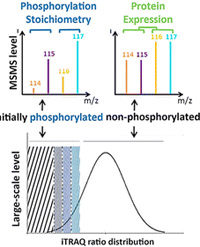
Phospho-iTRAQ: Assessing Isobaric Labels for the Large-Scale Study Of Phosphopeptide Stoichiometry
Pieter Glibert, Paulien Meert, Katleen Van Steendam, Filip Van Nieuwerburgh, Dieter De Coninck, Lennart Martens, Maarten Dhaenens, and Dieter Deforce
J. Proteome Res. (2015) 14: 839–849
With an estimated 30% of all proteins being phosphorylated, detailed analysis of the prevalence of this important modification is important for the elucidation of a range for cellular processes. While many workflows have focused on the enrichment of the phosphopeptides and subsequent quantitation by label-free or labeling approaches, the stoichiometry or occupancy of phosphorylation is not determined.
To measure the occupancy with the Phospho-iTRAQ method, a peptide sample is briefly split in two identical parts and differentially labeled preceding the phosphatase treatment of one part to remove the phosphate moieties. By focusing on the unmodified counterparts of phosphorylated peptides, the method circumvents the ionization, fragmentation, and enrichment difficulties that hamper quantitation of stoichiometry in most common phosphoproteomics methods. The technique was validated on multiple instrument platforms using a complex lysate of EGF-stimulated HeLa cells.
|
 |
 |
 |
 |
|
Mascot tip of the month
Computer hardware gets more powerful each year, so it’s a good idea to take advantage of this by migrating your Mascot Server and Mascot Distiller installations every few years. In both cases, you need to email support@matrixscience.com beforehand, to request a new product key. In the case of Mascot Server, you will probably want to preserve your search history, so that you can still load important result reports from the search log. This is easily done, and detailed migration procedures can be found here.
If possible, give the new server the same hostname as the old one. This will mean that links to past search results in Mascot Daemon will not break. If this is not feasible, and you want to repair these links, you can use an SQL command to update the information in the Daemon task database. For the default VistaDB engine, there is a table editor utility in the Daemon application directory and the procedure is described in the Daemon online help (Getting Started > Database Engines > VistaDB) |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|