|
To view this email as a web page, click here. |
 |
|
Welcome
Susan Weintraub and Marshall Pope are the distinguished guest speakers at our annual
Mascot Workshop and User Meeting
in St. Louis on May 31. More details are below - register to reserve your spot.
Using database search for site analysis of modifications is increasingly popular - possibly because its so easy!
This month's highlighted publication describes how to characterize low abundance cysteine modifications.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns glycopeptide analysis.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Site analysis
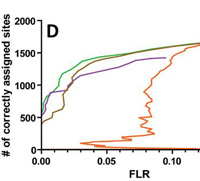
Were you aware that Mascot Server reports site assignments for modifications? Site analysis, which can be found in the Peptide View report (Mascot Server 2.4 and later) is based on the score differences between matches with different arrangements of modifications.
This approach was developed by Bernhard Kuster's group at TUM, who showed that the Mascot delta score outperforms other approaches by analysing a collection of synthetic analogs of real phosphopeptides.
Site analysis is available for any MS/MS spectrum that meets these requirements:
- The top scoring match contains one or more variable modifications for which alternative arrangements are possible.
- The top scoring match has a significant score.
- There is at least one further match to an alternative arrangement of modifications, which need not have a significant score.
If you would like to learn more about applying Site Analysis to your research, read
here. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Global Analysis of Myocardial Peptides Containing Cysteines With Irreversible Sulfinic and Sulfonic Acid Post-Translational Modifications
Jana Paulech, Kiersten A. Liddy, Kasper Engholm-Keller, Melanie Y. White and Stuart J. Cordwell
Molecular & Cellular Proteomics (2015) 14: 609-620.
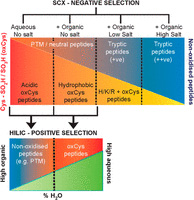
Cysteine oxidation is an important post-translational modification associated with redox signaling and oxidative stress. While there are a number of methods for the analysis of the reversible cysteine PTM's, the irreversible [-SO2H, -SO3H] modifications are more challenging. Not only is cysteine the second-least abundant amino acid in proteins, but these modifications occupy only 1-2% of these sites under physiological conditions, thus, efficient enrichment is critical.
The authors exploited the unique characteristics of tryptic peptides containing Cys-SO2H/SO3H to generate a novel method based on sequential negative selection by electrostatic repulsion using SCX followed by positive selection by hydrophilic interactions using HILIC. They identified 181 Cys-SO2H/SO3H sites from rat myocardial tissue subjected to physiologically relevant concentrations of H2O2 or to ischemia/re-perfusion injury.
|
 |
 |
 |
 |
|
Mascot tip of the month
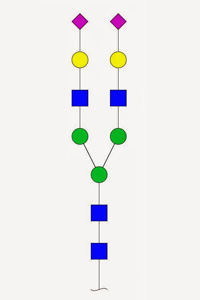
Glycopeptide analysis isn't easy. Many sugars are large and heterogeneous. In CID / HCD, the glycan tends to fragment at the expense of the peptide, so that sequence ions from the peptide are weak, and the entire glycan is lost, making site analysis impossible. ETD is preferred since it keeps the glycan intact and in situ.
Until recently, Unimod included a relatively small number of glycan entries, and none of these had the neutral losses defined, making it difficult to get decent matches using CID / HCD. This has been addressed, and the database now includes just about all naturally occurring glycans with masses of 2 kDa or less. The S and T specificities for O-linked glycans are grouped, so that they can be selected as a single modification, and all of them have neutral losses defined of zero, for ETD, and the complete glycan, for CID / HCD.
So, if glycopeptides are of interest, make sure you update the copy of Unimod on your in-house Mascot Server by downloading the latest unimod.xml. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|