|
To view this email as a web page, click here. |
 |
|
Welcome
What are the pros and cons of searching a database that is limited to specific proteins of interest?
In this month's featured publication, gel electrophoresis is combined with LC-MS/MS for complexome profiling.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month highlights a new video that illustrates how to configure a custom database using Database Manager.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Search only for peptides we care about?
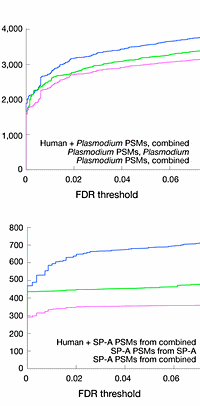
A recent note in Nature Methods by William Noble of the University of Washington proposes that researchers remove irrelevant peptides from the database prior to searching. He illustrates his point with a study of malaria, where the sample contained both human and plasmodium proteins. He observes that, if only the plasmodium proteins are of interest, you can get better sensitivity at a given FDR by removing the human sequences from the database.
This may be true for his example, but it is not a universal rule. If the spectra are 90% plasmodium and 10% human, searching a plasmodium-only database will almost certainly give better sensitivity. But, if the spectra are 10% plasmodium and 90% human, the sensitivity may get worse.
To read more about the potential benefits and limitations of searching a restricted database, please see this article. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Cryo-slicing Blue Native-Mass Spectrometry (csBN-MS), a Novel Technology for High Resolution Complexome Profiling
Catrin S. Mueller, Wolfgang Bildl, Alexander Haupt, Lars Ellenrieder, Thomas Becker, Carola Hunte, Bernd Fakler and Uwe Schulte
Molecular & Cellular Proteomics (2016), 15, 669-681
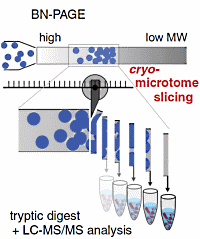
In this study the authors present a new high resolution workflow for characterization of native protein complexes. Using blue native (BN) gel electrophoresis combined with LC-MS/MS, it enables large scale identification of complexes and their subunits. This BN-MS workflow provides several advantages over previous approaches - it uses nearly the full resolving power of the BN-PAGE separation, it provides detailed abundance-mass profiles over a wide molecular size range, and it provides more comprehensive resolution of the subunit composition.
Sample preparation is performed by gel slicing with a cryo-microtome. For the analysis of a rat brain mitochondrial membrane preparation, it provided a total of 230 slices, each covering 0.3 mm of the BN gel. These gel slice samples were subsequently digested and analyzed by LC-MS/MS, and the data were evaluated using label-free quantification of proteins. In the entire set of cryo-slices, these analyses identified over 1,200 proteins, 774 of which were mitochondrial proteins.
Besides the higher resolution, the method was able to identify and quantify over 70% of the known mitochondrial proteome over the mass range of 100 to 3,500 kDa. |
 |
 |
 |
 |
|
Mascot tip of the month
How do you prefer to learn how to use new software? Web pages or a tutorial or muddle through and refer to reference material as needed?
It seems like the first choice for an increasing number of people is to watch a video. Recognising this, we've made the first of what we hope will be many videos, illustrating how to accomplish tasks in Mascot. Number one, now available on YouTube, deals with configuring a custom database using Database Manager.
We are very keen to hear your feedback on this initial offering. Too slow / too fast / too short / too long? Please let us know what works best for you and which topics you'd like to see covered. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|