|
To view this email as a web page, click here. |
 |
|
Welcome
When a digest is analysed by LC-MS/MS, the LC retention time contains valuable information. We discuss how this is handled within Mascot.
This month's highlighted publication presents a new database for protein complexes of plants, showing how co-migration can aid with identification.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains why summary reports no longer show non-significant matches by default, and
how you can revert this change if you wish.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Retention Time in Mascot
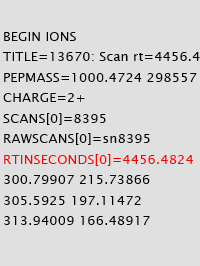
Though retention time is not utilized in Mascot scoring, it is increasingly used for re-scoring search results with Percolator as well as for many quantitation schemes. Mascot Server works off the peak list, not the raw data, so it is important to understand how retention time is transmitted through peak lists, search results and exports.
To ensure retention time could be explicitly and reliably included in an MGF peak list, we introduced the parameter RTINSECONDS. If the peak list is formatted as mzML, Mascot Server will look for the relevant controlled vocabulary (CV) terms.
When RT information is present in the MGF or mzML peak list, it will be output to the Mascot search result file. It can then be displayed in the Peptide View report, extracted using Mascot Parser, and exported in a variety of formats. If you are looking at results across several fractions, Distiller assigns an index to each raw file in the MGF header and references it at scan level.
One of the key advantages of making RT available is that it can be used by Percolator as a feature to improve sensitivity. Please go here to read more about how you can leverage retention time in your workflow. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
PCoM-DB Update: A Protein Co-Migration Database for Photosynthetic Organisms
Atsushi Takabayashi, Saeka Takabayashi, Kaori Takahashi, Mai Watanabe, Hiroko Uchida, Akio Murakami, Tomomichi Fujita, Masahiko Ikeuchi and Ayumi Tanaka
Plant Cell Physiol. (2016) online: December 22, 2016
To facilitate the understating of protein complexes in cellular processes, the authors have developed a searchable database of protein co-migration for photosynthetic organisms.
They used blue-native PAGE for separation, cut the gel strips into ~60 pieces, subjected them to in-gel digestion, and analyzed by LC-MS/MS with protein identification by Mascot. Protein abundances were estimated by spectral counting, using the exponentially modified protein abundance index (emPAI),
as implemented in Mascot.
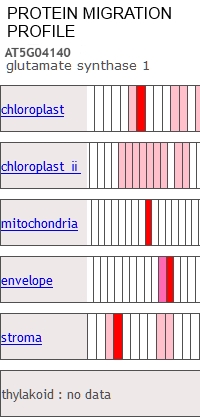
The Protein Co-Migration Database (PCoM-DB) provides prediction tools for protein complexes. PCoM-DB displays migration profiles for any given protein of interest, and allows users to compare them with migration profiles of other proteins, showing the oligomeric states of proteins and, thus, identifying potential interaction partners.
This paper covers the addition of data to the PCoM-DB from bryophytes, green algae, cyanobacteria, as well as Arabidopsis organelles, including intact chloroplasts, chloroplast stroma, the chloroplast envelope, and mitochondria.
|
 |
 |
 |
 |
|
Mascot tip of the month
In Mascot Server 2.6, we've made a small change that may have a big effect on the appearance of your search results.
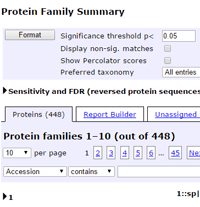
Previously, the installation default was to show all peptide matches in the body of the report, no matter how low the score. You could filter out the weak matches, if you wished, by entering a score or expect threshold value into the appropriate format control.
This worked fine for experienced users, but it was clear that some people, new to database searching, were assuming that all matches were good matches. This could be misleading, so we decided to change the installation default, and only display the significant matches. Instead of a control to enter a score or expect threshold, there is now a checkbox labelled Display non-significant matches. If you check this and choose Format, everything will be displayed, as before.
If you are a long-standing Mascot user, and prefer the old approach, you can revert this change. In the Configuration Editor, under Configuration Options, locate the parameter called DisplayNonSignificantMatches. Settings are:
- 0 = Display cleared check box in format controls (default)
- 1 = Display checked check box
- 2 = Display edit box for custom expect or score threshold
If you display the edit box, you can set a default value using IgnoreIonsScoreBelow. Otherwise, this setting is ignored. So, the old installation default corresponds to DisplayNonSignificantMatches 2 and IgnoreIonsScoreBelow 0.0 |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|