|
To view this email as a web page, click here. |
 |
|
Welcome
We will be exhibiting at the upcoming Proteomic Forum 2017, April 2-5 in Potsdam, Germany. Please stop by if you are attending.
We have some helpful suggestions if you are submitting search results to proteomics repositories such as PRIDE.
This month's highlighted publication demonstrates a new method for detecting residual proteins in plasma-derived immunoglobulin.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns processing options files in Mascot Distiller.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
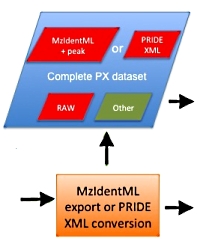
Submitting your search results to a proteomics repository
It is increasingly common for researchers to place their raw data and search results in a proteomics repository. Many journals now require this as a condition of publication.
A complete submission to the PRIDE repository consists of the raw data, the peak lists, and the search results in mzIdentML format. Here are a few tips to help you achieve a successful outcome.
- MIAPE compliance (see MIAPE-MSI for details)
- » Include protein description and length in the search result export to
enable protein sequence coverage in the mzIdentML file.
- » Run your searches in target/decoy mode so false discovery rates are included in the export file.
- Peak Lists
- » Always store a copy of the original peak list file in the same place as the mzIdentML file to ensure
accurate correlation between spectra and peptide matches.
- Modifications
- » Modifications not in the public Unimod database may be rejected by PRIDE.
Either re-run the search without the modification or add the modification to Unimod.
Go here to read about
these issues in greater detail. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
An application of mass spectrometry for quality control of biologicals: Highly sensitive profiling of plasma residuals in human plasma-derived immunoglobulin
Franck Limonier, Katleen Van Steendam, Genevieve Waeterloos, Koen Brusselmans, Myriam Sneyers, Dieter Deforce
Journal of Proteomics 152 (2017) 312-320
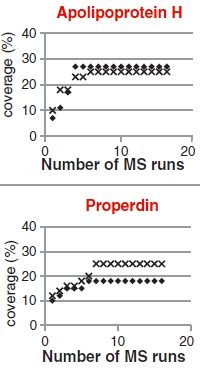
The authors have developed a mass spectrometry based method for the detection and quantitation of trace-level residual proteins in human plasma-derived immunoglobulin. These Ig's are used as therapies for disease-related immune deficiencies, as well as for autoimmune diseases. A series of adverse events due to these residuals is the stimulus for this work.
This study looked at a wide range of fractionation methods, acquisition strategies, replications, and samples to determine the optimum method. They concluded that enriching low abundant proteins using a bead-based combinatorial peptide ligand library (CPLL) is the preferred approach for detecting these residuals. Standard bottom-up LC/MS/MS and database search is used for detection and label-free quantitation.
They tested 5 different Ig products and discovered about 70 different residual plasma proteins. They demonstrated the quality control potential with a spiking experiment where levels of 1ng/mg in Ig were detectable, which is below the 3ng/mg threshold thrombotic dose (in vivo model).
|
 |
 |
 |
 |
|
Mascot tip of the month
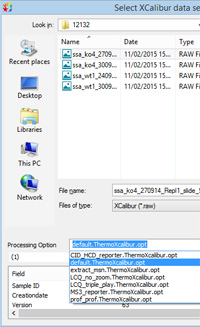
All the processing options files included with Mascot Distiller were revised for the 2.6 release. We also changed the location of these files and the way in which a file was associated with a particular raw file format. Just one problem - the path to the 'last used' options file in Distiller is sticky, so that when an old version of Distiller is updated, the first time you try to open an options file, it starts from the old location. It's possible that some people never realise that new files are available. Make sure you open and save options files in the new, default location (\ProgramData\Matrix Science\Mascot Distiller\processing options).
The recommended form for the name of an options file is exemplified by qtrap_reporter.SciexAnalyst.opt. Starting on the right, the extension must be opt. The next section determines the raw file format association, Sciex Analyst in this case. The rest of the name is free text, but if the text happens to be default then that file becomes the default for the associated format, e.g. default.WatersMS2E.opt.
This mechanism replaces the file associations dialog in older versions. If Distiller has been updated from 2.5 or earlier, you will still see old processing options files listed in the drop-down. We didn't want to delete old file associations automatically during the update in case they were in active use. If these old associations are simply cluttering up the list, you can easily remove them:
- Exit Distiller
- Open your Distiller settings file in a text editor: C:\Users\%USERNAME%\AppData\Roaming\Matrix Science\Mascot Distiller\Distiller.rst
- Delete all the lines between <Associations> and </Associations> (but don't delete these two lines)
- Save the changes
When you next launch Distiller, the drop-down will only contain new options files, as in the illustration on the right. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|