|
To view this email as a web page, click here. |
 |
|
Welcome
We have a suggestion for more efficient matching of trypsin autolysis peptides.
This month's highlighted publication shows a new way to determine kinase activity.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes how to disable result report caching.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Trypsin, trypsin, every where
Accurate identification of trypsin autolysis peptides is important. If the correct database matches are not available, these strong spectra can easily give rise to false positives. A recent paper from the Medical University of Graz pointed out that the enzyme usually employed in proteomics experiments is modified to inhibit autolysis. Unless the relevant modifications are included in a search, many of the autolysis peptides will go unmatched.
Problem is, there are many modifications. Typically, you might need Methyl (N-term), Methyl (K), Dimethyl (K), Dimethyl (N-term), Dehydro (C), Deamidated (NQ) and Carbamidomethyl (N-term) as variable mods. In the sample we studied, we also had to use semiTrypsin as the enzyme, because it was clear that there were large numbers of non-specific autolysis peptides.
At 1% peptide FDR, the counts of PSMs and distinct sequences for the semi-tryptic search with multiple varmods were 545 and 49 compared with 281 and 10 for a search with strict trypsin and Carbamidomethyl (N-term) as the only variable mod.
This is good for trypsin but bad for the proteins of interest. No-one wants to burden their searches with such a large search space. The solution is to create a custom spectral library. By including this in the search, you can use tight search parameters yet still obtain matches to all the the weird and wonderful trypsin autolysis peptides, removing them as a possible source of false positives. This article describes the procedure to create such a custom spectral library.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
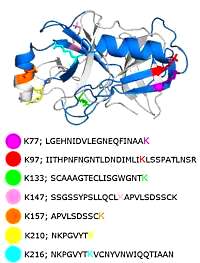
Kinase activity ranking using phosphoproteomics data (KARP) quantifies the contribution of protein kinases to the regulation of cell viability
Edmund H. Wilkes, Pedro Casado, Vinothini Rajeeve and Pedro R. Cutillas
Molecular & Cellular Proteomics, 2017, 16, 1694-1704
The authors set out to develop a model for the contribution of protein kinases to cell viability. Their starting premise is that the fraction of protein phosphorylation attributed to a given kinase relative to total phosphorylation may be a measure of its contribution to signaling.
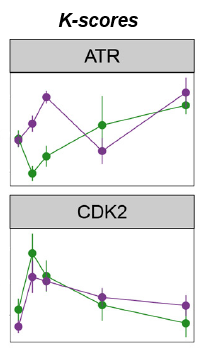
Using LC/MS/MS datasets of phosphopeptide enriched, digested cell lysates, they developed the algorithm, which calculated the sum of the intensities of its known substrates relative to the sum of the intensities of all phosphorylation sites present in the dataset, producing the K-score.
To investigate whether the K-scores reflected the contribution of kinases to cell viability, and could therefore predict responses to compounds, they measured reductions in cell viability in a panel of cell lines as a function of treatment with nine kinase inhibitors. They found a significant correlation between K-scores of given kinases and reduction of cell viability after their inhibition.
|
 |
 |
 |
 |
|
Mascot tip of the month
The use of cache files to speed up loading of result reports was introduced in Mascot Server 2.3. The files are created the first time a summary report is loaded, and speed things up when the same report is loaded subsequently. Cache files also speed up the loading of peptide view and protein view reports.
Naturally, there is some overhead to creating these cache files. If you rarely open a report more than once, and rarely look at peptide or protein view reports, you might save time by disabling caching.
To disable caching for a single summary report, just add &_usecache=0 to the URL.
To disable caching globally, in mascot.dat, look for the two lines beginning ResfileCache and ResultsCache. Comment them out, and add new lines containing just these keywords. That is, your mascot.dat will look like this
# ResfileCache master_results.pl, master_results_2.pl, peptide_view.pl, protein_view.pl, export_dat.pl, export_dat_2.pl, ms-createpip.exe, MSAnatomiser.class, mi_getpeaklist.pl, msms_gif.pl, nph-mascot.exe, ms-searchcontrol.exe, client.pl
# ResultsCache master_results.pl, master_results_2.pl, peptide_view.pl, protein_view.pl, export_dat.pl, export_dat_2.pl, MSAnatomiser.class, mi_getpeaklist.pl, nph-mascot.exe, ms-searchcontrol.exe
ResfileCache
ResultsCache
This change will increase memory usage by some scripts and utilities; best to ensure you have at least 16GB of RAM.
It will also make changing pages or tabs or expanding sections of the protein family summary slower. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|