|
To view this email as a web page, click here. |
 |
|
Welcome
Did you realise that instances of Mascot Daemon running on different PCs can share the same task database, so that everyone sees the same set of tasks? More details in our first article.
This month's highlighted publication describes a detailed approach to discovering and validating peptide biomarkers for ovarian cancer.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains why it is often worth searching a UniProt reference proteome rather than just setting a taxonomy filter on SwissProt.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
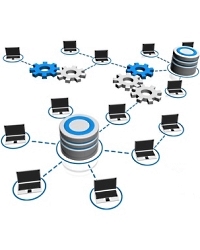
Sharing the Daemon task database
If you have a busy lab with automated searches being submitted from different locations and instruments, it may be helpful to centralize the database that controls these tasks. By default, each copy of Daemon uses its own task database to store information about the tasks it is running and links to the results. Sharing the task database allows you to track all the searches from all locations, although each task is still 'owned' by the Daemon instance that launched it, and only the owner can pause or cancel the task.
If you are using a file-based database engine, such as VistaDB or Access, you will need to switch to one of the server-based options: MySQL, PostgreSQL, SQL Server, or Oracle. Daemon includes an import/export facility that make migration easy.
Additionally, one of the new features in Mascot Daemon 2.6 is the ability to set a task to start on the completion of another task. You can then chain a set of tasks to run in a custom order, task 2 on completion of task 1, task 3 on completion of task 2 etc. Tasks can be scheduled first come, first served or in another order that reflects the importance or urgency of the job.
Go here to read more details about setting up and using a shared task database with Mascot Daemon.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Selected reaction monitoring approach for validating peptide biomarkers
Qing Wang, Ming Zhang, Tyler Tomita, Joshua T. Vogelstein, Shibin Zhou, Nickolas Papadopoulos, Kenneth W. Kinzler, and Bert Vogelstein
PNAS 2017 114 (51) 13519-13524; published online December 4, 2017
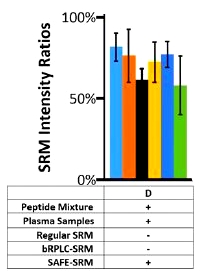
The authors have developed a selected reaction monitoring (SRM)-based method for the discovery and validation of peptide biomarkers for ovarian cancer. It utilizes multiple steps of fractionation and separation as well as synthesis of target peptides, and they call it "sequential analysis of fractionated eluates" - SAFE-SRM.
The study was executed in three phases
- Phase 1, global proteomic profiling of plasma samples from cancer patients and healthy individuals, yielding 641 candidate peptide markers from 188 genes
- Phase 2, implementation of a SRM-based assay to evaluate each of the 641 candidate peptide markers in additional plasma samples, yielding two peptides from peptidyl-prolyl
cis-trans isomerase A as promising biomarkers
- Phase 3, evaluation of the performance of these two peptides in an independent set of cancer patients and controls using SAFE-SRM.
To provide validation a separate cohort of 73 cases, consisting of plasma from 35 ovarian cancer cases and 38 samples from healthy individuals or patients with other cancer types, was tested against the two PPIA peptides. Twenty (57.1%; 95% CI, 40-73%) of the 35 plasma samples from ovarian cancer cases scored positive for one or both of these peptides. None of the 14 samples from normal individuals scored positive.
|
 |
 |
 |
 |
|
Mascot tip of the month
For a number of good reasons, SwissProt is a very popular choice for general purpose searching. But, if you are working on a well characterised organism, you are likely to get better coverage from a UniProt reference proteome.
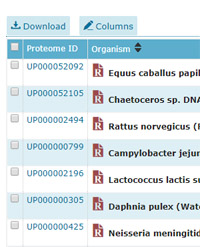
Taking the obvious example of Human, there are 20,245 entries with a taxonomy of Homo sapiens in the current SwissProt compared with 71,785 proteins in the reference proteome. The additional 51k entries have been selected from the 141k human entries in Trembl, and some information about the selection criteria can be found here. Clearly, if the additional sequences are well chosen, we can expect a worthwhile increase in coverage.
To illustrate, the human proteome was downloaded using the UniProt proteome template in Database Manager. This includes isoforms, which increases the entry count to 93,799. Searching a typical human data set (HEK 293 data from Pride project PXD001468) against SwissProt returned 102,206 distinct sequences at 1% FDR while the search of the reference proteome using identical settings returned 105,140 distinct sequences - almost 3k additional sequences.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|