|
To view this email as a web page, click here. |
 |
|
Welcome
We are looking forward to seeing many of you at the upcoming World HUPO meeting in Florida, where we are offering free advice sessions.
We analyse the speed increases for peak picking and quantitation of profile data with Distiller 2.7.
This month's highlighted publication shows a method for determining protease substrates in extracellular matrix.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how and why we filter out matches to short peptides.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Free consultations at World HUPO
- Have a question about error tolerant searches or post-translational modifications?
- Thinking of trying a new quantitation method?
- Want to add a new database to Mascot Server or set up Mascot security?
- Need advice on how to interpret your search results?
We are offering free advice on the best use of Mascot Server, Mascot Daemon and Mascot Distiller at the 17th World HUPO Conference in Orlando, Florida (30 September to 3 October).
Thirty minute time slots are available for a maximum of three people at a time. There won't be time to run long searches or process large raw files so, once you have booked a slot, you are welcome to send us data in advance. If you wish to do this, contact support@matrixscience.com to discuss what files to send.
Location will be the Matrix Science exhibition booth, which is #307. To book a slot, follow this link.
|
 |
 |
 |
 |
|
Faster peak detection with Mascot Distiller 2.7
The latest version of Mascot Distiller has significantly improved peak detection speed due to changes in the handling of profile data. In previous versions, peak picking required that the profile data was on a linear mass scale, with uniformly spaced values. In many cases, this meant that it was necessary to interpolate large numbers of additional data points into each scan. In Mascot Distiller 2.7, peak detection works directly on the raw profile data, without the need to re-grid.
To evaluate the change we took a SILAC quantitation dataset from a Thermo QExactive comprised of 10 files, with a total of 1.2 million MS and MS/MS spectra. We then processed them with Distiller 2.6 and 2.7. Peak picking time dropped from 8 days to 3 hours and quantitation from 2.5 days to 16 hours.
In addition to greatly speeding up the processing time, we found approximately 20% more significant peptide matches at a 1% FDR.
Go here to read more about these improvements.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
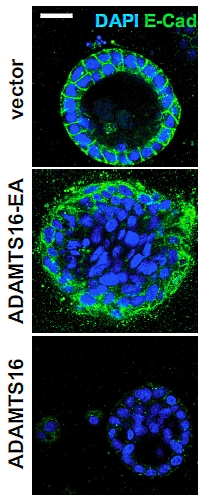
A Selective Extracellular Matrix Proteomics Approach Identifies Fibronectin Proteolysis by A Disintegrin-like and Metalloprotease Domain with Thrombospondin Type 1 Motifs (ADAMTS16) and Its Impact on Spheroid Morphogenesis
Rahel Schnellmann, Ragna Sack, Daniel Hess, Douglas S. Annis, Deane F. Mosher, Suneel S. Apte, and Ruth Chiquet-Ehrismann.
Molecular & Cellular Proteomics, 2018, 17, 1410-1425
The authors investigated the role of the secreted protease ADAMTS16 and its effect on fibronectin (FN) and the assembly of the extra cellular matrix (ECM). Until now ADAMTS16 had been an orphan protease, without known substrates, although it has been linked to several human diseases such as cancer and hypertension.
They used a cell-free ECM produced in vitro by BALB/c fibroblasts and seeded it with HEK-EBNA cells that were transfected to produce the ADAMTS16 protease. HEK-EBNA cells stably expressing various ADAMTS16 constructs were seeded on top of the FN and the medium was analyzed by Western blotting and LC-MS/MS.
The authors confirmed a large reduction of FN fibrils in the presence of ADAMTS16, and temporal analysis of the FN assembly suggests that ADAMTS16 primarily interferes with initiation and maturation of the fibrils. Additionally the release of the 30 kDa FN fragment indicates that ADAMTS16 is also capable of degrading FN fibrils.
|
 |
 |
 |
 |
|
Mascot Tip
There are two settings in mascot.dat that control the handling of very short peptides. Any peptide sequence from the database shorter than MinPepLenInSearch is simply ignored. Any peptide sequence match shorter than MinPepLenInPepSummary is never treated as sufficient evidence, by itself, for the presence of a protein. That is, if you had MinPepLenInSearch set to 7 and MinPepLenInPepSummary set to 10, you would see matches to 7, 8, and 9-mers in the reports but you would never see a protein split out as a new family or new family member on the basis of one of these matches. Each family or family member would have to contain a significant match to a peptide that was at least a 10-mer and wasn't found in any other family or family member.
In recent versions of Mascot, both cut-offs are set to 7 on installation. Please resist the temptation to reduce either to a low value. It is difficult or impossible to get a significant match to a very short peptide because the probability that the sequence could occur in the database by chance is too high. Setting a low value for MinPepLenInSearch will have an impact on search speed and the size of the result file. Setting a low value for MinPepLenInPepSummary can cause catastrophic over-clustering in the protein family report (because very short sequences occur by chance in unrelated proteins).
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|