|
To view this email as a web page, click here. |
 |
|
Welcome
We hope to see some of you at the Proteomic Forum in Potsdam, Germany later this month.
Neutral losses can be leveraged to identify very labile modifications, such as O-glycosylation.
This month's highlighted publication introduces a new bead-based method for extracting and identifying proteins from cell lysates.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to describe a problem when requesting technical support.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
O-glycosylated peptides: neutral loss to the rescue
Since the O-glycosidic bond is weaker than the peptide backbone, typical MS/MS conditions cause it to break, leaving behind mostly unmodified fragments. A recent paper on the characterization of O-fucosylated thrombospondin type 1 repeats from malaria parasites highlighted the difficulty of identifying these modified peptides.
By correctly configuring the neutral loss definitions for these modifications, O-fucosylation can readily be identified with Mascot. If a variable modification has multiple neutral losses, all are tried during the database search. The one giving the highest score is chosen and the rest are treated as satellite peaks.
If you find that likely neutral losses are missing from a definition, such as C-mannosylated tryptophan losing 120 Da (C4H8O4), these are easy to add using the Mascot Server Configuration Editor.
We demonstrate the utility of Mascot for identifying these O-glycosylated peptides in the search of a recombinant P. falciparum thrombospondin data set. Go here to read the details of the analysis.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Proteomic Sample Preparation through Extraction by Unspecific Adsorption on Silica Beads for ArgC-like Digestion
Yannik Lewin, Moritz Neupartl, Vahid Golghalyani, and Michael Karas
J. Proteome Res., 18 1289−1298 (2019)
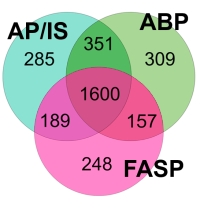
The authors have introduced an adsorption-based protocol (ABP) for the extraction of proteins from cell lysates. The method uses noncovalent immobilization on paramagnetic silica particles. This also enables the modification and tagging of protein samples under harsh conditions, such as organic solvents, high salt concentrations, or low pH values. Additionally the method reduces sample preparation time by 40-60% compared with acetone precipitation followed by an in-solution digestion (AP/IS) or filter aided sample preparation (FASP). Protein identifications are also slightly increased compared to AP/IS or FASP.
They then combined the ABP approach with ArgC-like digestion, which uses trypsin after propionylation of lysines, causing an inhibition of one of the cleavage sites of trypsin. The number of identifications achieved by the ArgC-like protocols showed an 18% increase over those from standard tryptic digestion. In comparison to trypsin they also observed lower occurrences of missed cleavages in peptides, cleaner fragmentation in favor of y-ion series, and a disproportionately higher number of identifications of membrane proteins and histones.
|
 |
 |
 |
 |
|
Mascot Tip
Some software issues are easy to describe - maybe the full text of the error message is all that is required. For Mascot Server, you can copy and paste error messages from the Mascot Server error log (errorlog.txt in the Mascot logs directory). If you are working with client software, and want to ask about a message box that has popped up, did you know that you can capture the text to the Windows clipboard by using CTRL+C while the message box is active? This is usually more useful than a screen shot and a lot easier than trying to type the text into an email.
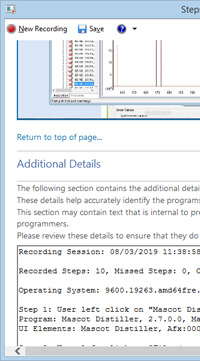
Other problems with client software can be more difficult to describe. Maybe multiple steps are involved, or it's a case where 'a picture is worth a thousand words'. This is where the Windows Problem Steps Recorder can be enormously helpful. This tool is available in Windows 7 onwards, but is somewhat hidden away. You can launch it by typing psr.exe in a command window or in the search bar. It records what's happening on the screen and saves a report in a zipped MHTML file, which is ideal for sending as an email attachment. The report automatically includes screen shots, timings, and software versions. You can even highlight areas of the screen and add comments as you go along.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|