|
To view this email as a web page, click here. |
 |
|
Welcome
Please join us for our annual ASMS User Meetings in Atlanta on June 3rd and 4th.
We have some suggestions for configuring an X!Hunter MGF format spectral library.
This month's highlighted publication shows how Mascot can be used to identify polymeric contaminants in your peptide samples.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes the benefits of consolidating Mascot Server licences.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Matrix Science ASMS User Meetings
Join us for breakfast and learn about exciting applications of database searching as well as how to get the most out of Mascot.
There is no charge for attending these meetings, but advance registration is required. Breakfast will be provided.
Monday 3 June, 7:00 am - 8:00 am
Database searching in systems without a genome: lessons learned with the California sea lion
presented by Ben Neely, Marine Biochemical Sciences Group, NIST
Tuesday 4 June, 7:00 am - 8:00 am
A day in the life of a database search engine: Mascot as robust and scalable tool for protein identification in a contract research environment
presented by Michael Ford, MS Bioworks
|
 |
 |
 |
 |
|
Using the X!Hunter MGF spectral library
Mascot 2.6 gives you the ability to use a variety of formats for your spectral library searching - the NIST MSP format, the SpectraST sptxt format and the X!Hunter MGF format. X!Hunter, though minimally annotated, offers over 200 discrete libraries from guinea pig and cow to Brucella and Shigella.
You can readily set up a single-file library using Mascot Server Database Manager, as we illustrate for the M. tuberculosis H37Rv MGF library. Searching a whole-cell lysate of M. tuberculosis against this library produced 17,444 hits out of the 65,878 queries. However, we chose to use a more stringent qualifier for higher confidence and reduced this to 1,641 strong peptide matches and 256 protein families.
If you want a human or mouse library, the files are organized by chromosome, so the 20 or so separate files need to be downloaded. To utilize these files requires them to be concatenated into a single library file; otherwise the setup is the same as for a single file.
Go here to read more details about setting up an X!Hunter MGF spectral library.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
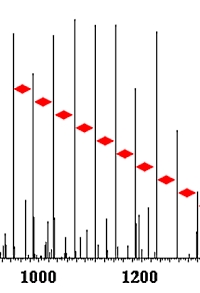
Identification of Poly(ethylene glycol) and Poly(ethylene glycol)-Based Detergents Using Peptide Search Engines
Shiva Ahmadi and Dominic Winter
Anal. Chem., 90 6594-6600 (2018)
The authors have developed a method for detecting and identifying the very common polyethylene glycol (PEG) contaminants that plague many protein samples. The approach is to identify PEG and PEG-based detergents using peptide search engines that utilize an artificial amino acid in combination with a custom-made database.
The letter J was defined as the artificial amino acid C2H4O, representing the ethylene glycol monomer unit. A PEG database was then designed consisting of PEG molecules containing 1 to 100 monomers. Since the PEG detergents contain a variety of head groups, variable modifications to J were included, corresponding to the common detergents Tergitol, Tween20, Tween80, Triton X-100, Triton X-114, Brij35, and Brij58.
Upon LC-MS analysis of contaminated in-gel digested fractions from B104 cell secretome, 806 PEG spectra originating from four PEG species were identified. Using error-tolerant and mass-tolerant searches resulted in the identification of 3409 and 3187 PEG-related MS/MS spectra, respectively.
|
 |
 |
 |
 |
|
Mascot Tip
It isn't unusual to have multiple Mascot Server licenses in the same laboratory. Maybe they came with different instruments or maybe it is the result of some lab reorganisation. In most cases, there are real advantages to consolidating the licences into a single, larger licence:
- The opportunity to get greater speed for individual searches
- Only one server to administer and keep the software and sequence databases up-to-date
- All search results in one place
If there are multiple groups of users in the lab, who need to keep their databases or search results private, this can be handled by Mascot security.
Best of all, you save money. A 2 CPU licence costs less than 2 x 1 CPU licences, and this extends to the cost of annual support or a version update. We don't charge to create a merged licence, but the systems must be on the same version. If one is earlier than the other, you'll need to buy a version update for the older one. The other requirement is that you have suitable hardware. Either a single PC with sufficient cores or, for cluster mode, PCs with identical hardware.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|