|
To view this email as a web page, click here. |
 |
|
Welcome
Get an insight into Mascot scoring statistics and how to optimize your searches.
This month's highlighted publication shows a new method to identify epitopes using direct infusion.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month looks at choosing between Windows and Linux as the operating system for your Mascot Server.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
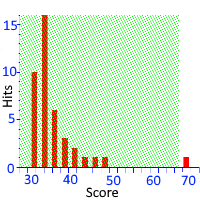
Unravelling Mascot's Statistics
Mascot uses probability-based scoring to determine whether a match is significant or not. That is, it calculates the probability that the observed match is a chance (or random) event. When we report the scores, we use the negative log of this probability so that, when the probability of a random match is very small and highly unlikely, the reported score will be high.
The important question is where and how the line is drawn between scores that are significant and scores that represent chance matches. This depends on a number of assumptions. The key modeling assumption is that all of the peptide sequences within precursor tolerance are random, so-called true nulls. Obviously this is not strictly true in a real search, where the database is chosen with the goal that the majority of spectra have at least one "correct" sequence within precursor tolerance. The model is robust and handles this well as long as the proportion of true nulls remains high.
Another assumption is that the sequences are statistically independent. This isn't always true. A real database contains highly homologous proteins, and it's possible to get clusters of peptide sequences that only differ by a small mutation. PSM scores for these peptides will be correlated, which can skew the distribution of p-values within a query.
Go here to read more about how multiple peptide matches are handled, and the importance of using a target/decoy search to validate the calculated statistics.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
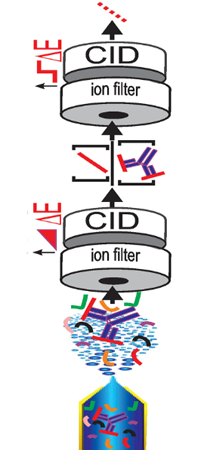
Intact Transition Epitope Mapping - Targeted High-Energy Rupture of Extracted Epitopes (ITEM-THREE)
Bright D. Danquah, Claudia Rower, Kwabena F. M. Opuni, Reham El-Kased, David Frommholz, Harald Illges, Cornelia Koy and Michael O. Glocker
Molecular & Cellular Proteomics, 18 1543-1555 (2019)
The authors have developed a method for identifying epitope peptides using direct nano-electrospray of antigen/antibody solutions. The process involves trypsin digestions of the antigen, 1-hour incubation of these peptides with the antibody, then directly spraying this unseparated solution into the MS.
The crux of the method involves first filtering out the unbound peptides with an m/z 5000 cutoff at the first quadrupole, collisionally dissociating the bound peptides from the antibody, separating these peptides from the antibody in the mobility cell, then fragmenting the peptides, and recording their spectrum with the TOF analyzer.
They validated the method using mixtures of epitope-containing synthetic peptides incubated with antiTRIM21 and antiRA33 antibodies. Further work identified two previously unknown epitope peptides from the enzymatic digest of recombinant human TNFα and an antiTNFα antibody.
|
 |
 |
 |
 |
|
Mascot Tip
We are often asked whether it is better to choose the Windows or Linux editions of Mascot Server. Our standard reply is that you should choose the one that the system administrator prefers. Someone needs to be responsible for the server, if only to make sure the system doesn't grind to a halt because the disk is full. While most Mascot housekeeping is handled through the web interface, there are certain tasks that require a knowledge of the OS, such as applying software patches. No good choosing Linux if no-one in the group has any experience of working at a Linux command-line.
Operating system cost is sometimes a factor. Many Linux distributions are available for free, as long as you don't require technical support. Windows is not free, but Mascot Server does not require Windows Server, and will operate perfectly happily under something like Windows 10 Professional, although there are certain limits that could be a problem for a busy server with many users. For example, recent PC versions of Windows have a limit of 20 concurrent TCP/IP connections. If multiple users have multiple browser windows open to the server, plus a few file sharing connections, this limit can soon be reached. There are also limits on hardware, but these are pretty generous. For example, Windows 10 Professional supports 2 processor sockets, 256 logical cores, and 512GB RAM.
In terms of Mascot Server performance and functionality, there is very little to choose. Mascot Daemon and Mascot Distiller are Windows-only, but there is no requirement to run these on the same machine as Mascot Server. On the other hand, if you regularly submit very large searches from Mascot Daemon, running Daemon on the Mascot Server desktop allows it to bypass the 4GB upload limit of the Microsoft web server, which could be an important factor in your choice.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|