|
To view this email as a web page, click here. |
 |
|
Welcome
Mascot Distiller gains a new capability: ion mobility data can now be used to improve quantitation.
This month's highlighted publication shows a quantitative study of melanoma and a potential prognostic model.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot Server 2.8 has been released!
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
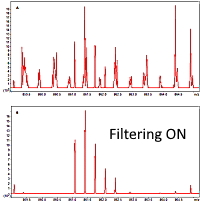
Ion mobility filtering improves quantitation
The latest version of Mascot Distiller utilizes the ion mobility data generated by the Bruker timsTOF instrument to improve the precision of precursor-based quantitation methods, such as SILAC or Label-Free.
Distiller uses the precursor ion mobility values for identified peptides as a filter on the precursor scans during the XIC peak detection steps. This cleans up and improves the quality of the data in the target precursor regions by removing interfering signals as shown on the right.
For a given peptide match, an ion mobility range is applied using the precursor ion mobility grouping tolerance defined in the processing options. Only MS1 signals from within the calculated ion mobility range are then used during XIC peak detection.
For a HeLa protein digest spiked with an E. coli digest, quantification was substantially faster (10 vs 36 hrs.) with the ion mobility filtering. Additionally, median peptide fraction and correlation values also improved, allowing more peptide ratios to pass the quantitation quality thresholds.
Go here to read more about enabling ion mobility filtering.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Proteomics of Primary Uveal Melanoma: Insights into Metastasis and Protein Biomarkers
Geeng-Fu Jang, Jack S Crabb, Bo Hu, Belinda Willard, Helen Kalirai, Arun D Singh, Sarah E Coupland, John W Crabb
Cancers 2021, 13, 3520
The authors pursued quantitative proteomic analysis for insights into the mechanisms of uveal melanoma (UM), the most common primary malignancy of the eye. They investigated pathways of metastasis and discovered biomarkers for protein-based methods of UM prognosis.
Using 8-plex iTRAQ LC-MS/MS quantitation they identified and quantified 3935 proteins with 2 or more unique peptides, with the proteomes of the metastasizing and non-metastasizing UM appearing about 89% similar. Comparison of the average protein ratios from metastasizing and non-metastasizing UM yielded 402 differentially expressed (DE) proteins that contained less than 20% missing data and a minimum fold-change of one standard deviation. Of these, 191 proteins were more abundant in metastasizing UM and 211 proteins were more abundant in non-metastasizing UM.
Pathway analysis of the DE proteins elevated in metastasizing UM predicted significant over-representation of immune system pathways, possible targets for immune checkpoint therapy. The pathway analysis of the DE proteins elevated in non-metastasizing UM showed over-representation of the pathways involving metabolism.
The authors also developed a statistical model for prediction of UM metastasis using 32 proteins; 17 proteins were positively correlated with metastasis, and 15 negatively correlated. This model provided an area under the ROC curve of 0.93, sensitivity of 0.91 and specificity of 0.81.
|
 |
 |
 |
 |
|
Release of Mascot Server 2.8
Version 2.8 of Mascot Server was released in July, and there are new features that you may find very helpful in your work. If you were under warranty or support in July, you were emailed with download information for your free update. If this message was missed for some reason, please contact us to obtain your update.
Important new features include:
- New statistical model for error tolerant searches. Mascot assigns expect values to ET matches and (if the search is auto-decoy) estimates FDR.
- Increased Percolator sensitivity. Mascot ships with a new Percolator feature set, and the Percolator version has been updated.
- MS/MS searches are faster. Singly threaded file processing steps have been multithreaded, yielding 20-35% speed improvement.
More information about these new features on this page.
If you plan to move to new hardware at the same time as updating to 2.8, consult our step-by-step migration procedures.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|