|
To view this email as a web page, click here. |
 |
|
Welcome
Peptide fragmentation spectra from single-cell samples present some new challenges for database searching.
This month's highlighted publication describes a method to use Volumetric Absorptive Microsampling (VAMS) devices for rapid trypsin digestion.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
We have new pricing for updating older Mascot Server licences.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
Challenges of protein and peptide identification with single cells
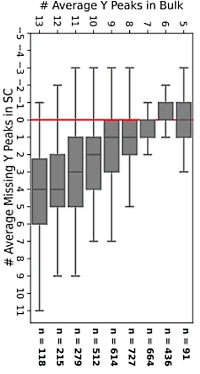
Instrumentation and sample preparation for single-cell proteomics is improving. Investigating the heterogeneity of single cells presents some challenges to database searching, as all algorithms, including Mascot, have been developed using bulk samples from organisms, tissues and cells.
Intensity compression can blur the line between signal and noise. When Mascot matches theoretical peaks to the MS/MS spectrum, it divides the spectrum into 100Da bins and looks for fragment ions in each bin. As long as the noise is random and doesn't completely overwhelm the fragment peaks in every bin, the algorithm is usually able to pick the signal from the noise. Adding noise tends to decrease the match score rather than prevent a match.
Fewer fragment peaks could be observed in single-cell MS/MS spectra compared to bulk spectra. A recent computational study compared the number of y fragments and also showed that single-cell spectra tend to be quantitively distinct from bulk spectra. If you use spectral libraries, these should be made from single-cell data, not bulk spectra.
Score thresholds may be affected, since the scores of candidate peptides matched to the spectrum can be lower due to intensity compression and fewer fragment peaks. Re-scoring matches with a machine learning tool like Percolator is worth a try to improve sensitivity.
More details in our blog, including an example Mascot search of a single-cell digest.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Next generation VAMS®-Trypsin immobilization for instant proteolysis in bottom-up protein determination
Léon Reubsaet, Bernd Thiede, Trine Grønhaug Halvorsen
Advances in Sample Preparation, 3 (2022) 100027
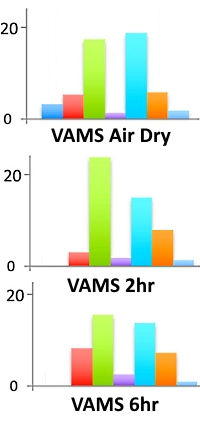
Based on their experience with immobilized trypsin on paper-based samplers, the authors investigated the use of immobilized trypsin on Volumetric Absorptive Microsampling (VAMS) devices for preparation of samples for LC/MS. They optimized trypsin immobilization, looking at the effect of pH, reaction time, volume and concentration of the enzyme. Optimal values were found to be pH=9 and 50µg of trypsin.
Using a 7-protein equimolar mixture, they evaluated the time dependence of the digestion with VAMS. The workflow captures the proteins on the derivatized VAMS tips, then either lets them dry (and digest) immediately, or the tips are kept moist in a closed container to allow them to continue reacting with the trypsin for several hours. The VAMS trypsin samplers then undergo post-digestion reduction and alkylation before peptide extraction for LC/MS. Signal intensities for the tryptic peptides were similar for the rapid digestion (air dried) and for the 2-hour and 6-hour tests.
Using VAMS for a serum sample with air drying provided the identification of 96 proteins, while the overnight VAMS digestion yielded 99 proteins. For comparison, an in-solution digest of the sample gave 204 proteins.
|
 |
 |
 |
 |
|
New pricing for updating older Mascot Server licences
We are reducing the price for updating Mascot Server 2.4 and earlier licences. The cost to update Mascot to the latest version is determined by how many versions behind your licence is. One-version update is 25%, 2-version 50% and 3-version update 75% of the cost a new licence. From 11 July 2022, the 4-version update is fixed at 90% of the cost of a new licence. This means you now get a 10% price reduction when you trade in Mascot 2.4 or older for 2.8, no matter how old your licence is.
This is an excellent time to update. Mascot 2.8 not only supports the latest versions of Windows and Linux, but all updates come with one year of Premium Support. This includes priority technical support by e-mail and phone and remote problem diagnosis if necessary. If you have any trouble updating Mascot or migrating to new hardware, we are here to help you.
More details about update pricing are on our website, including links to features and improvements since Mascot 2.5. Please contact us to request a quote or if you have any questions.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|