|
To view this email as a web page, click here. |
 |
|
Welcome
There is updated guidance from the Human Proteome Project for mass spectrometry data interpretation.
This month's highlighted publication shows a new method for the chemical labelling and quantification of cysteine containing peptides and proteins.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month offers assistance with creating new crosslinking methods.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
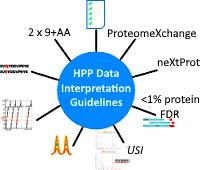
Human Proteome Project data interpretation guidelines update
HUPO's Human Proteome Project recently updated their guidelines for MS data interpretation. While the focus is human, many of the guidelines are good practice and common sense in any proteomics study. Here are some of the key updates.
- Deposit all MS proteomics data to a ProteomeXchange repository as a complete submission
- Include analysis reference files (search database, spectral library, transition list, etc.) in the submission
- Use the most recent version of the neXtProt reference proteome
- Describe in detail the calculation of FDRs at the PSM, peptide, and protein levels
- Report the PSM-, peptide-, and protein-level FDR values along with the total number of expected false positives at each level
- In DDA with high confidence ID's, consider alternate mappings of the peptides to proteins other than the claimed one
Go here to read about the latest guidance in greater detail.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
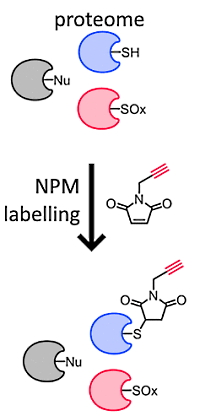
Maleimide-Based Chemical Proteomics for Quantitative Analysis of Cysteine Reactivity
Evan W. McConnell, Amanda L. Smythers, and Leslie M. Hicks
J. Am. Soc. Mass Spectrom. 2020, published online: June 23, 2020
The authors have developed a method to quantify site-specific cysteine reactivity using a maleimide thiol-reactive probe (N-propargylmaleimide, NPM.) Called QTRP (quantitative thiol-reactivity profiling) the method involves cysteine labelling with NPM followed by either linking a fluorescence probe (for gel-based analysis) or biotin (for affinity purification and LC/MS/MS) via the alkyne moiety.
The LC-MS/MS approach used streptavidin-biotin enrichment with on-resin proteolytic digestion. Bound peptides were cleaved from the resin using formic acid, leaving a unique NPM-DADPS modification (+278 Da) on labeled Cys residues that can be identified by database searching. Applying this method to the proteome of the unicellular green alga Chlamydomonas reinhardtii, they quantified 1585 Cys-sites across 870 proteins.
|
 |
 |
 |
 |
|
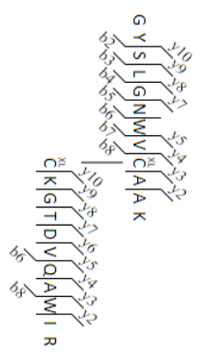
Mascot Tip
Have you tried matching crosslinked peptides in Mascot Server 2.7? We've seen it give some very interesting results, especially for natural crosslinks, like disulfide bridges and isopeptide bonds.
Providing the flexibility to handle real life data leads to some unavoidable complexity. For example, Lysine can form an isopeptide bond with Glutamine but you don't expect bonds between two Lysines or two Glutamines. Spelling this out is the job of the crosslinking method. We try our best to provide clear documentation for configuring a new method, with realistic examples, but there can still be a steep learning curve.
Getting started is much less of a challenge if you have a good model on hand, where reasonable choices have been made for the settings. You can immediately see how things are supposed to work and focus on optimisation.
We are happy to offer assistance with this. Contact support with some basic information about the experiment. We will send an upload link to our dropbox. Send us one raw file, (no more - we don't need a huge data set to illustrate suitable settings). We will return a custom crosslinking method together with recommendations for search parameters and peak picking.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|