|
To view this email as a web page, click here. |
 |
|
Welcome
For some environments, single sign-on can enhance security and ease login fatigue.
This month's highlighted publication examines zirconium and titanium magnetic particles for phosphopeptide capture.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns parameters for handling variable modifications.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Securing Mascot with Single Sign-On (SSO)
Mascot Server is often shared between individuals or groups who require different levels of security and privacy for their searches. Although Mascot Security provides the mechanism to limit access to result reports, databases, and even custom modifications, a determined hacker could probably find ways around it without too much difficulty.
If the security is a concern, we recommend keeping your Mascot server behind a firewall or restricting access with a strong authentication system. One approach is to use a single sign-on (SSO) system, where you only need to authenticate yourself once to gain access to a range of applications and software systems. Mascot Server is compatible with these systems as it has no direct integration with authentication systems; all the heavy lifting is done by the web server.
The web browser sends information about the user to the web server, which then will interact with an authentication system. If authentication succeeds, the web server will pass the name of the authenticated user to the Mascot Security user database. If the user exists, they are "logged in" to Mascot without prompting for a password.
You must ensure that the Mascot user name matches the external user name, and the Mascot user must be configured to use web server authentication. Go here to read more details about setting up the SSO.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Zirconium(IV)-IMAC Revisited: Improved Performance and Phosphoproteome Coverage by Magnetic Microparticles for Phosphopeptide Affinity Enrichment
Ignacio Arribas Diez, Ireshyn Govender, Previn Naicker, Stoyan Stoychev, Justin Jordaan, and Ole N. Jensen
J. Proteome Res. 2021 20(1) 453-462
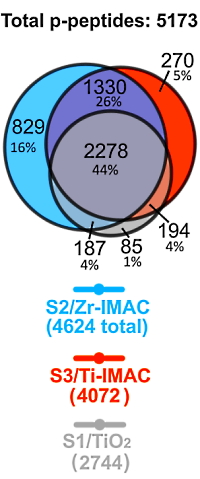
The authors investigated the impact of solvent conditions on the enrichment of phosphopeptides in protein digests. They compared Zr-IMAC magnetic microparticles to Ti-IMAC and TiO2 under 6 different hydroxy acid solutions and characterized the retained peptides by LC-MS/MS.
The best-performing enrichment methods (Solvent2/Zr-IMAC, Solvent3/Ti-IMAC, and Solvent1/ TiO2) were then further assessed by automated phosphopeptide enrichment and LC−MS/MS of the tryptic digest of human HepG2/C3A cells. With the optimized conditions the Zr-IMAC retrieved more phosphopeptides (4624) than Ti-IMAC (4072) or TiO2 (2744).
In total, 5173 phosphorylated peptides were identified by the combination of S2/Zr-IMAC, S3/Ti-IMAC, and S1/TiO2. Almost half of these phosphopeptides (44%) were recovered by all three methods (2278). S2/Zr-IMAC retrieved the highest number (829) of method-unique phosphopeptides, 16% of the total.
They then compared these methods with the standard LC-based Fe-IMAC approach. While the total number of phosphopeptides identified was similar, the enrichments performed with magnetic microparticles were much more selective with more than 90% of the total peptides assigned as phosphopeptides, while the selectivity of Fe-IMAC HPLC was 78%.
|
 |
 |
 |
 |
|
Mascot Tip
Handling of variable modifications became more sophisticated in Mascot Server 2.7. Three parameters are used to tune for greater speed or greater depth:
- MaxPepNumVarMods is the maximum number of different variable modifications allowed on an individual peptide. The default is 3. But, if you are studying hyper-modified proteins, like histones, or very long peptides, you can increase this value at the expense of search speed.
- MaxPepNumModifiedSites is the maximum number of modified residues per peptide. Similar considerations as for MaxPepNumVarMods apply to changing the default value of 5.
- MaxPepModArrangements is the maximum number of arrangements of an individual varmod composition. If site analysis is not important in your identifications, you might wish to reduce the default value of 64.
Examples of how these parameters influence search results can be found in a recent blog article.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|