|
To view this email as a web page, click here. |
 |
|
Welcome
Identifying endogenous peptides is easier with recent improvements to Mascot.
This month's highlighted publication shows how get better peptide recoveries from solid phase extraction.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month has some housekeeping advice concerning the Mascot Daemon task database.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Identify more endogenous peptides
Identifying endogenous peptides is challenging since a no-enzyme search uses a very large search space, and so score thresholds tend to be conservative and sensitivity is reduced. The results can usually be improved by re-scoring the database matches with Percolator to improve separation between true and false matches.
Using datasets from a library of 2000 synthetic HLA peptides recently published, we compared the performance of the new 2.8 version of Mascot with the previous version. A major addition to 2.8 is the enhanced Percolator functionality which now encompasses 46 features, as well as improved handling of retention times.
Using Percolator in Mascot 2.7 improves the target PSM count by 2.5x at 1% PSM FDR and also increases the number of identified peptide sequences by 75%. But Mascot 2.8 with Percolator now gives a 3.6x increase in target PSMs and over 2x increase in identified peptides.
The results can be further improved by enabling the retention time feature in Percolator. Using the RT feature in Mascot 2.7 made little difference in the number of PSMs or identifications, where with 2.8, it gives an additional ~14% boost in target PSMs and 6% increase in identified peptides.
Go here to read more about improving your searches.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
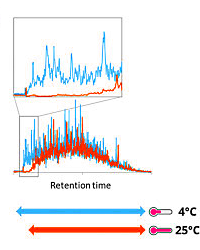
CoolTip: Low-Temperature Solid-Phase Extraction Microcolumn for Capturing Hydrophilic Peptides and Phosphopeptides
Kosuke Ogata and Yasushi Ishihama
Molecular & Cellular Proteomics, published online: November 02, 2021
Off-line solid phase extraction (SPE) using pipette tips containing various reversed-phase resins is widely used for capturing peptides due to the large capacity, high recovery, and low elution volume required. However, hydrophilic peptides such as very short peptides and phosphorylated peptides are often not well retained in the solid phase and are easily lost during the purification process resulting in insufficient proteome coverage.
Using tryptic peptides from HeLa cell lysates, the authors investigated various solvent and media systems, concluding that all combinations provided similar modest recovery of hydrophilic peptides. They then evaluated the SPE tips at various temperatures (25 °C, 16 °C, 8 °C, and 4 °C) and found that the recovery of hydrophilic peptides increased as the tip temperature was lower.
They confirmed that this "CoolTip" protocol (when compared with the room temperature method) yielded about 5% more peptide identifications in the analysis of the entire lysate, while the phosphoproteomics-focused experiments yielded about 15% more. They assessed the CoolTip protocol using label-free quantification and found the recoveries of peptides and phosphopeptides at elution time < 20 min. were approximately doubled.
|
 |
 |
 |
 |
|
Mascot Tip
If you make heavy use of Mascot Daemon, you probably rely on the Daemon task database for organising and locating past search results. Losing the database through disk failure or corruption can be very inconvenient, even though the search results are still present on the Mascot Server. Hopefully, as with any important document, you make regular backups. If you use the default VistaDB database engine, here is a tip to minimise potential disruption.
VistaDB is a simple database that stores the information in a single file. To avoid this file becoming unwieldy, archive the file and start a new one every so often:
- In Daemon preferences, on the ODBC connection tab, make a note of the path to the database file. The default is C:\ProgramData\Matrix Science\Mascot Daemon\TaskDB.vdb5
- Exit the Daemon desktop application then stop the Daemon engine. This may be running in the system tray (right click the icon and choose exit) or as a Windows service (Windows control panel; Administrative tools; Services; Mascot Daemon Engine; Select; Stop)
- In Windows task manager, verify there is no running process called MascotDaemon.exe or MascotDaemonEngine.exe
- Rename the database file to (for example) TaskDB_2021.vdb5
- Replace it with a copy of the empty database file supplied in the VistaDB sub-directory
- Re-start Mascot Daemon
If you want to work with an archived task database file, just enter the filename in the ODBC connection tab and choose Save. When you are done, revert to the current file.
You can also use this procedure in the event of the task database file becoming damaged or lost. If you need help with trying to recover a damaged file, email support@matrixscience.com describing the problem. You can attach the file if it is less than a couple of MB. Otherwise, use a dropbox to send it to us.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|