|
To view this email as a web page, click here. |
 |
|
Welcome
With the common FTP protocol fast disappearing, you may need to review your download methods.
This month's highlighted publication shows a middle-down method to quantify histone PTMs and their crosstalk in malaria parasites.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month contains a reminder about a recent Macot Distiller update
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Retiring the File Transfer Protocol (FTP)
Downloading protein sequence databases via Mascot Server Database Manager has usually been handled with the File Transfer Protocol or FTP, but after 50 years of service, this protocol is heading for retirement. Support for FTP has been removed from common browser platforms as well as from NextProt.
HTTPS is increasingly used as the default protocol, and all the major protein sequence databases are available via FTP and HTTPS, including NCBI, EBI and Uniprot, with some allowing HTTP. One of the main advantages of HTTPS over FTP is the built-in error detection, which FTP lacks. If there are any transfer errors, such as an unreliable network or data packets arriving in the wrong order, the connection terminates with an error. The HTTPS client can then resume the download from the last known good position.
As of November 2021, Database Manager uses HTTP (HTTPS if necessary) for commonly used databases like SwissProt, contaminants and all the EST databases, as well as taxonomy files downloaded from NCBI. If you have an older version of Mascot Server please read here about steps you need to take to ensure your downloads work properly.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
A dynamic and combinatorial histone code drives malaria parasite asexual and sexual development
Hilde von Gruening, Mariel Coradin, Mariel R. Mendoza, Janette Reader, Simone Sidoli, Benjamin A. Garcia, Lyn-Marie Birkholtz
Molecular & Cellular Proteomics, published online: 17 January 2022
The authors investigated the histone code of the malaria parasite Plasmodium falciparum in order to understand the nature of the PTMs that act in various levels of coordination. Since peptide-centric proteomics experiments tend to produce short peptides that often do not span multiple PTMs, they adopted a middle-down method to produce longer reads and more readily see the relative quantities of the various PTMs.
They cultivated the P. falciparum and isolated parasites as enriched trophozoites, immature early-stage gametocytes, and mature gametocytes to determine stage-specific differences. Digestion using GluC produced peptides ~50-60 AA in length, which were then analyzed by LC-MS/MS using ETD fragmentation. After searching against PlasmoDB, they further removed any ambiguously mapped PTMs. Using IsoScale they determined the relative quantification of co-fragmented isobaric peptides that share the same sequence but have distinct positions of the PTMs.
77 PTMs on histones H3 and H3.3 were identified and quantified, with 34 described for the first time. The stage-specific analysis showed the influence between pairs of PTMs, and they quantified their co-existing frequency as an "interplay score". A third of the co-existing PTMs had similar frequencies across the three development stages while several PTMs show an interplay score that is uniquely associated with a specific parasite life cycle stage. For example, in trophozoites, the majority of the PTM combinations had negative interplay scores, suggesting that these PTMs antagonize each other, and are thus mutually exclusive.
|
 |
 |
 |
 |
|
Mascot Tip
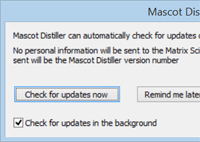
The current release of Mascot Distiller, version 2.8.1, includes several important bug fixes. We recommend installing this update. Note that Distiller updates are free.
If you are using an earlier version, and haven't been prompted to update, this may be because you have turned off update reminders. To re-enable update checks, from the Help menu, choose Check for updates... then tick the option to Check for updates in the background. Or, in version 2.8, choose a suitable interval from the drop-down list.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|