|
To view this email as a web page, click here. |
 |
|
Welcome
MS/MS spectra of crosslinked peptides are inherently complex. We looked at the effects of peak picking parameters for getting best results.
This month's highlighted publication shows a new approach for targeted protein dephosphorylation.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot Distiller supports generating HTML and CSV reports for quantitation results on the command line.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
Best peak picking for intact crosslink spectra
Crosslinking studies provide valuable information on interaction sites between proteins, but the resulting MS/MS spectra are often very complex, with overlapping contributions from both linked peptides. Peak picking these spectra can be challenging, so we investigated the effects of several Mascot Distiller parameters on the resultant peak lists.
Crosslink spectra often have higher charge state precursors and fragments, so these must be de-charged before database searching. Alternatively, export the fragment charge state in the MGF file (supported by Mascot Server 2.7 and later). If your MS/MS scans are saved as centroids, rather than profile, then you need to use the uncentroiding options before Distiller can determine charge states.
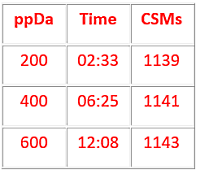
For test data, we used a publicly available synthetic peptide library with known crosslinks. We tried peak picking by 1) taking the centroid values from the raw file without determining charge state; 2) uncentroiding the MS/MS scans at a resolution of 100 points per Dalton (ppDa) and de-charging; and 3) uncentroiding the scans at 600 ppDa and de-charging.
These 3 methods yielded 655, 1087 and 1143 significant cross-linked spectrum matches (CSMs), respectively. The de-charged peak lists also give increased residue coverage because the higher charge state fragments can now be matched, improving the score.
Additionally, the uncentroiding resolution (ppDa) was varied from 600 to 200 to see the effect on the resulting number of CSMs. The number of CSMs reduced a little while the processing time decreased by a factor of 5. To read more suggestions on improving your peak picking of crosslinked spectra, please go to the
blog post.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
An affinity-directed phosphatase, AdPhosphatase, system for targeted protein dephosphorylation
Luke M. Simpson, Luke J. Fulcher, Gajanan Sathe, Abigail Brewer, Jin-Feng Zhao, Daniel R. Squair, Jennifer Crooks, Melanie Wightman, Nicola T. Wood, Robert Gourlay, Joby Varghese, Renata F. Soares, and Gopal P. Sapkota
Cell Chemical Biology 30, 188–202, February 16, 2023
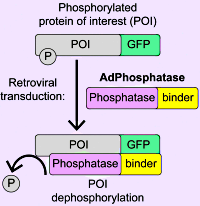
The authors have developed a new approach termed AdPhosphatase, which efficiently and selectively targets GFP-tagged proteins for dephosphorylation. The AdPhosphatase system consists of a binding nanobody conjugated to the catalytic subunit of a phosphatase (PPP1CA/PPP2CA).
They compared the efficacy of the AdPhosphatase-directed dephosphorylation to that of a small molecule kinase inhibitor and found a comparable reduction in phosphorylation between the two approaches.
Using TMT-labeled quantitation and phosphopeptide enrichment and analysis, 11,821 unique phosphopeptides were identified. Of these, levels of phosphopeptides corresponding to 21 proteins were significantly downregulated by more than 2-fold by the AdPhosphatase. Additionally, the AdPhosphatase-mediated dephosphorylation was remarkably specific, with only a small list of non-targeted phospho-peptides shown to be significantly downregulated.
|
 |
 |
 |
 |
|
Batch scripting with Distiller quantitation reports
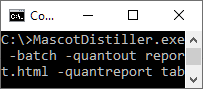
Mascot Distiller is predominantly a GUI application, but some of the Quantitation Toolbox functionality can be used on the command line. One use case is creating a quantitation report for an existing project that already contains quantitation results. The command line is:
MascotDistiller.exe project.rov -batch -quantout output.html -quantreport table-peptides.py -logfile output.log
The syntax and arguments are briefly described in the Distiller help, Reference, Command line arguments. The first part, -batch -quantout file.html, triggers a quantitation report, and the -quantreport table-peptides.py argument names the Python script that implements the report. The default output format is HTML. You can pass additional parameters depending on the Python script, and the available parameters can be found in the associated XML file, for example Mascot Distiller\reports\table-peptides.py.xml. Table-peptides supports output format CSV:
MascotDistiller.exe project.rov -batch -quantout output.csv -quantreport table-peptides.py -exportFormat CSV -logfile output.log
The 'batch' argument is so named due to historical reasons, and it simply means don't start the GUI. Only one .rov file is processed even in batch mode. The above command is quite useful when you have searched and quantitated a raw file through Mascot Daemon using Distiller as a data import filter. Under external processes, add the quantreport call in section "After completing task" with suitable arguments, and you'll automatically get a CSV report at the end of the Daemon task.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|