|
To view this email as a web page, click here. |
 |
|
Welcome to issue #101
We are looking forward to seeing many of you at the upcoming ASMS meeting in Houston, Texas, where we will host our Annual User Meeting. Please also stop by at exhibit booth #726.
This month’s highlighted publication shows the negative effect of trace metal ions on quantifying phosphopeptides and what to do about it.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
We are also celebrating the 25th anniversary since the founding of Matrix Science. Coincidentally, the March 2023 newsletter was the 100th monthly issue.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
Matrix Science at ASMS
We are pleased to invite you to our Annual ASMS User Meeting in Houston, Texas on Monday, 5 June at 7:00am – 8:00am, which will feature the following presentations.
- Special guest presentation – Professor Susan Weintraub, University of Texas Health Science Center at San Antonio
- Single cell proteomics with Mascot – Patrick Emery, Matrix Science
- 25 years of Matrix Science – Ville Koskinen, Matrix Science
There is no charge for attending these meetings, but advance registration is required. Breakfast will be provided. Please
register on our website.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
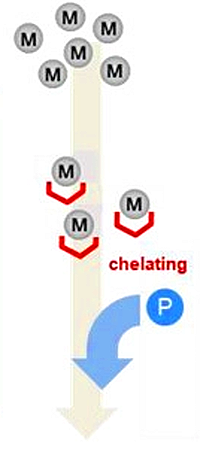
Bioinertization of nanoLC/MS/MS systems by depleting metal ions from the mobile phases for phosphoproteomics
Yumi Komori, Tomoya Niinae, Koshi Imami, Jun Yanagibayashi, Kenichi Yasunaga, Shinya Imamura, Masami Tomita, Yasushi Ishihama
Molecular & Cellular Proteomics, in press, 2023
The authors have developed a method to improve the analysis of phosphopeptides by minimizing the distortion of peak shape and reduction of signal intensity that occurs when metal ions are present. The method also avoids some of the limitations of additives that can suppress ionization and cause unwanted precipitation.
Approximately 3,600 diverse phosphopeptides were used as model substrates to investigate the effects of metal ions on quantification of protein phosphorylation in trace samples. Phosphopeptides were enriched from a HeLa cell extract/digest using hydroxy acid-modified metal oxide chromatography with TiO2 particles. The authors tested the adsorption of this mixture on the common chromatography materials of stainless steel, titanium, and PEEK and found recovery of multiply-phosphorylated peptides to be 4.3- and 5.4-fold decreased for stainless and titanium, while the recovery from PEEK was near 100%. When using solvent that had passed through the pump and mixer to dissolve the sample, they found significant suppression from metal ions, while solvent from before the pump did not have this effect.
They examined off-line chelating resin columns and found them effective for metal ion depletion. This led to the development of an on-line metal depletion system using chelating resin columns prepared by inserting an Empore iminodiacetate-immobilized membrane disk into a PEEK union and placing it between the nanoLC pump and injector. Metal-free PEEK and fused silica capillaries were used downstream of the on-line device. The peak areas of phosphopeptides were increased by an average of 9.9-fold with the on-line metal ion removal device, while for multiply-phosphorylated peptides, peak areas increased an average of 77-fold.
|
 |
 |
 |
 |
|
25 years of Matrix Science
Matrix Science is 25 years old. A lot has happened since John Cottrell and David Creasy founded the company in 1998, and proteomics today certainly looks very different to the bleeding edge of science back then.
From the humble beginnings when Matrix Science licensed the MOWSE III (Molecular Weight Search) algorithm, developed by Darryl Pappin and David Perkins (Imperial Cancer Research Fund, UK), the Mascot search engine is now cited in over 2000 papers every year. Thousands of people trust Mascot, and there are millions and millions of Mascot search results in existence.
The success of any company could be attributed to various causes, but the simple fact is that we would not be here without our loyal customers. Thank you for your trust, custom, bug reports, ideas and feedback over the past quarter century.
You can read more about our history and the evolution of the technology and our products in
our recent blog.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot. Read more about the company on our about page.
|



|
 |
|
|