|
To view this email as a web page, click here. |
|
|
|
Welcome
We have new recommended settings for Proteome Discoverer and Mascot Server 3.1 when you enable machine learning.
In this month's highlighted publication, mitochondrial acetylation is explored using an engineered acetyltransferase.
We are pleased to announce a new Mascot distributor in India.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Machine learning, MudPIT scoring and Proteome Discoverer
|
|
|
Mascot Server 3.1 includes functionality to enable predicted intensities (MS2PIP) and predicted retention time (DeepLC) through Proteome Discoverer.
The trick is to create a custom instrument definition in the Mascot configuration editor, then select that instrument in PD.
Enabling predictions almost always yields more peptide matches, but we were puzzled why it did not increase the number of quantified proteins.
The solution turns out to be a setting in the Mascot node in PD.
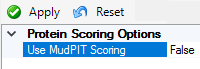
Mascot search results have two protein scoring algorithms, Standard and MudPIT.
MudPIT has been the default for a very long time (since Mascot Server 2.0 in 2004), and it's also the default in PD.
However, the calculation is not appropriate for protein FDR thresholding when machine learning is enabled.
Try switching from MudPIT to Standard scoring in PD settings and reprocess the file.
When machine learning is enabled, Mascot returns a logarithm of the PEP (posterior error probability) as the peptide match score.
Standard scoring means that PD will simply sum the best log(PEP) for each peptide, leading to a statistically valid protein score.
Now the Protein FDR Validator node is able to correctly set protein FDR.
We processed a benchmarking LFQ data set with MudPIT and Standard scoring.
Enabling predictions yielded 8% more peptide matches in this data set, compared to just using the Percolator node.
Changing to Standard scoring translated this into 5% more quantified proteins.
The details are in our blog.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Mitochondrial hyper-acetylation induced by an engineered acetyltransferase promotes cellular senescence
Tadahiro Shimazu, Ayane Kataoka, Takehiro Suzuki, Naoshi Dohmae, Yoichi Shinkai
iScience, Volume 28, Issue 9113233 (2025), doi:10.1016/j.isci.2025.113233
The NAD-dependent protein deacetylase SIRT3 is localised in the mitochondria and is responsible for mitochondrial deacetylation.
The authors have developed an engineered mitochondrial acetyltransferase (eMAT), which induces global lysine acetylation (AcK) in mitochondria.
The goal is to evaluate lysine hyper-acetylation beyond SIRT3 knockout models.
Characterisation, localisation and function of eMAT were explored with immunofluorescense microscopy, flow cytometry and LC-MS/MS.
The authors also found that eMAT promotes cellular senescence in normal human diploid h-TERT RPE1 cells.
To identify proteins acetylated by eMAT, the authors performed label-free quantitation and SILAC experiments using Mascot Server and Proteome Discoverer.
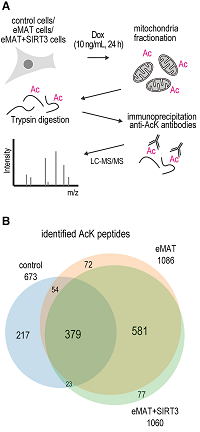
Mitochondria were isolated from control (HEK293T), eMAT-treated and eMAT+SIRT3 treated cells, digested with trypsin and peptides immunoprecipitated with anti-AcK antibodies.
After LC-MS/MS, 1403 peptides were identified in total.
The overlap (88%) of identified peptides between eMAT and eMAT+SIRT3 implies that SIRT3 can target proteins acetylated by eMAT.
Multiple acetylation sites on metabolic enzymes were identified, with their acetylation levels controlled by eMAT and SIRT3.
|
|
|
|
 |
|
|
|
New Indian distributor for Mascot
|
|
|
We are pleased to announce a new distributor for Matrix Science products!
Customers in India and neighbouring countries can now buy Mascot Server through BioInnovations, who provide instruments, software, services and consumables for life sciences and analytical and clinical labs.
In addition to India, the distribution agreement covers Bangladesh, Bhutan, Maldives, Nepal and Sri Lanka.
BioInnovations are a Bruker channel partner and provide MALDI and timsTOF instruments for proteomics.
Bruker BioTools and BioPharma Compass include native integration with Mascot Server for trusted protein identification by peptide mass fingerprinting and LC-MS/MS.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|





|
|
|
|
|