|
To view this email as a web page, click here. |
|
|
|
Welcome
We are developing a new spectrum-centric scoring algorithm for peptide identification from DIA spectra.
This month's featured publication uses Mascot Server and Distiller to identify and localize phosphorylations, leading to a proposal for a novel splicing regulation mechanism.
A recent update for Windows 11 and Server 2025 can cause issues with Mascot and the IIS webserver.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Spectrum-centric scoring for DIA
|
|
|
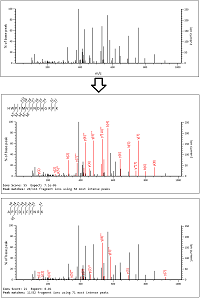
Data-Independent Acquisition (DIA) mass spectrometry often suffers from an overemphasis on maximizing protein identification numbers at the expense of the quality of results.
To address this, we're working on a spectrum-centric solution, called Mascot DIA.
Unlike conventional peptide-centric methods, the goal is to explain all the observed peaks in the MS/MS scan, without prior assumptions about peak intensities or elution times.
The core algorithm is noise-resistant scoring.
Mascot compares the theoretical fragments, including neutral losses, against the observed spectrum, and calculates the probability that it is a chance match.
To achieve a statistically significant score, a match must demonstrate sufficient, non-random sequence coverage.
Mascot DIA will not have to deconvolute spectra along the time axis or create pseudo-DDA projections or use predicted spectra from a library.
High mass accuracy and peak intensities are used when selecting peaks, and the noise model has been redesigned to better account for peaks from unrelated peptides.
Probabilistic scoring ensures that only sufficiently well-fragmented peptides are identified while limiting the number of false positives.
More details and a SCIEX SWATH example are available in our blog.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Mass Spectrometry and 3D Modeling Indicate the SBK2 Kinase Phosphorylates Splicing Factor SRSF7 to Regulate Cardiac Development
Mark Bouska, Eduardo Callegari, Daniela Paz and Xuejun Wang
Kinases Phosphatases 2025, 3(4), 20, doi:10.3390/kinasesphosphatases3040020
The study delivers the first in vivo evidence that SH3 Domain Binding Kinase Family Member 2 (SBK2) plays a vital role in left ventricular function.
The authors studied restoring the levels of SBK2 in mouse models and used mass spectrometry and AlphaFold structural modeling to identify its partners and elucidate the mechanisms of action.
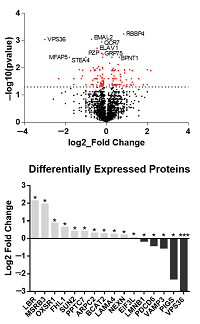
Quantitative proteomics was used to identify up and down-regulated proteins in SBK2 overexpression in mouse ventricles.
Pathway analysis showed SBK2 as an agent in actin/myosin cytoskeletal organization and localized in the PTW/PP1 phosphatase complex.
This led to phosphorylation enrichment studies using a dual enrichment method.
Mascot Server and Distiller were used to identify and localize the phosphorylation using the built-in site analysis algorithm.
Mascot identified RABX5/RABGEF1 as well as Serine and Arginine Rich Splicing Factor 7 (SRSF7) as the top phosphorylated proteins.
RABGEF1 is a ubiquitin-binding protein that targets mitochondria and is known to cause severe mitochondrial dysfunction, while SRSF7 is known to regulate the transition from a juvenile to adult-type transcriptome.
The phosphorylation site data was combined with computational analysis and modeling to determine the interaction between SBK2 and SRSF7.
This led the authors to propose a novel mechanism by which SBK2 phosphorylates SRSF7 and regulates the splicing factor to promote cardiomyocyte development.
|
|
|
|
 |
|
|
|
Windows update issue with IIS
|
|
|
The recent KB5070773 update to Microsoft Windows 11 and Windows Server 2025 can cause issues with the IIS webserver, preventing Mascot Server and the IIS welcome page from being viewed.
This includes access to the webserver when using 'localhost', computer name and IP address.
The problem may not manifest itself immediately, and it depends on a variety of conditions including other updates, Internet connection and computer restarts.
If you are experiencing this issue, make sure all updates have been installed, then reboot the computer.
Rebooting should initiate an automatic "Known Issue Rollback" that resolves it.
For organisations with managed devices, the IT department will need to implement a special Group Policy to deploy the rollback.
Once the issue has been resolved, Mascot Server will work again without any lasting damage.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|





|
|
|
|
|