|
To view this email as a web page, click here. |
|
|
|
Welcome
Mascot Daemon includes a summary report for top-3 quantitation, illustrated here with DDA-PASEF data.
This month's highlighted publication provides ZooMS peptide markers for armadillos, anteaters and sloths.
Migrating from Windows 10 to Linux can extend the lifetime of your hardware.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Top-3 quantitation using DDA-PASEF
|
|
|
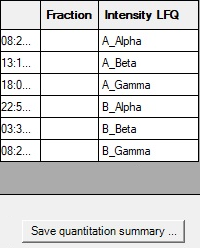
Mascot Daemon includes a protein level quantitation summary report for both precursor based methods (LFQ, SILAC etc) and MS/MS quantitation (iTRAQ, TMT).
Additionally, Daemon 3.0 supports top-3 ("Average") quantitation, where the three most intense peptides are averaged.
Top-3 quantitation is supported with all DDA instruments.
To illustrate, we selected raw files from PRIDE project PXD028735 acquired using a Bruker timsTOF (DDA-PASEF).
This project has two hybrid proteome samples (A and B), each with three replicates, comprising of known quantities of human, yeast and E. coli proteins.
The raw files were processed through Daemon using the default timsTOF Mascot Distiller processing options and with precursor ion mobility filtering enabled.
At 1% peptide sequence FDR, a total of 41,068 peptide sequences were identified from 372,444 significant PSMs, mapping to 8,001 proteins.
We defined a simple sample map for the quantitation summary using unique_mz, meaning the intensities for different charge and modification states of the same peptide are not summed together.
Then, click a button to generate a CSV file, which can be loaded into Excel or R.
The expected log2 ratios for human, yeast and E. coli were 0, 1 and -2, while top-3 quantitation reported -0.117 (±0.191), 0.725 (±0.238) and -1.819 (±0.362).
Median average deviation is lower compared to using total intensity, as the top-3 method is less likely to be influenced by outliers.
The processing steps, screenshots and results are available in our blog.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Peptide Mass Fingerprinting of South American Xenarthrans: A New Resource for Zooarcheology and Palaeontology
Mariya Antonosyan, Roshan Paladugu, Michael Ziegler, Gabriela Prestes Carneiro, Eliane Chim, Andre Menezes Strauss, Diego Mendes, Rafael Lemos, Jorge Domingo Carrillo-Briceño, Laura Pereira Furquim, Stefanie Schirmer, Jana Ilgner, Daniela Volke, Patrick Roberts
J. Proteome Res., November 3, 2025, doi:10.1021/acs.jproteome.5c00636
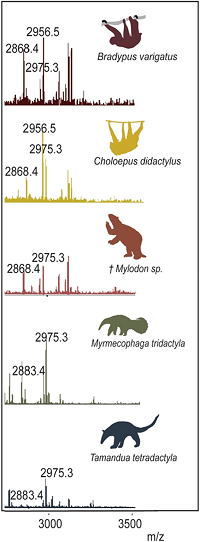
The authors report a unique set of collagen type I (COL1) peptide markers for differentiating South American Xenarthra species, including armadillos, anteaters and sloths, which are under-represented in reference databases.
The goal is to enable taxonomic identification of fragmented and morphologically indistinct bone samples using peptide mass fingerprinting (zooarcheology by mass spectrometry, ZooMS).
Reference bone samples of extant and extinct sloths and armadillos were assembled from the Museum für Naturkunde (Leibniz Institute for Evolution and Biodiversity Science), Anthropological Museum of the Federal University of Goiás, Laboratory of Archeology Curt Nimuendaju and Zoological Museum of the University of Zurich.
Publicly accessible annotated genomes of the extant taxa and one extinct species were digested in silico to identify suitable candidate marker sequences for COL1.
After a tryptic digest, the samples were analysed with MALDI-TOF (Bruker AutoFlex), yielding a presence/absence matrix of 12 standard markers.
The candidates were then confirmed using LC-MS/MS (Orbitrap Exploris 480 and Waters Synapt).
The protein sequences of the representative model organism in each taxonomic group were combined with mammalian COL1 orthologs.
After a tryptic digest, the spectra were searched with Mascot error tolerant search to allow for sequence variations and unsuspected variable modifications.
The authors were able to confirm peptide markers for ten extant and extinct species, confirming the current phylogenetic framework of Xenarthra.
|
|
|
|
 |
|
|
|
Migrating from Windows 10 to Linux
|
|
|
Microsoft ended Windows 10 mainstream support on 14 October 2025.
If your PC hardware is still in good condition but isn't compatible with Windows 11, there's no need to throw it away.
Mascot Server is compatible with all recent Linux distributions.
There is no functional difference between the Windows and Linux version, although we recommend some familiarity with Linux administration if you intend to switch.
If you have an active support contract, switching is included free of charge; please contact us for advice.
If you have an earlier Mascot Server 2.x licence, we recommend updating to the latest version.
A version update comes with 1 year of Premium Support, including a free platform switch and guided installation and migration.
Version 2.x is eligible for a 50% discount regardless of the number of CPUs and age of the licence.
Please contact us for more details and a free quote.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|





|
|
|
|
|