|
To view this email as a web page, click here. |
|
|
|
Welcome
Proteomics software is prone to filling up your disk, which can cause strange failures.
This month's highlighted publication characterises proteomic signatures in human fingertip regeneration.
The column widths in the Mascot search log can be customised using a little-known configuration option.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Strange software failures? Check your disk space
|
|
|
From time to time, a piece of software that has been working reliably suddenly doesn't.
The most common reason for Mascot Server to fail is running out of disk space, which can cause all kinds of weird faults.
Proteomics software is more prone to it than others due to the large size of input data and temporary files created during processing.
We've collected tips and advice for diagnosing and fixing disk space issues in our blog.
For Mascot Server, the three largest consumers of disk space are sequence databases ('sequence' directory), results files for database searches ('data' directory) and cache files created for reports ('data/cache' directory).
Mascot Server ships with a tool, tidy_data.pl, for compressing old search results and deleting unused cache files.
With Mascot Distiller, the largest consumers of disk space are raw files, the project files (.rov) and temporary files created during normal operation.
If your disk is full of .rov files, then it's safe to move them to a different disk.
Just make sure you also move the raw files to the same disk as the project files contain links to the raw data.
Distiller also includes a menu action to delete unused cache files.
Finally, Mascot Daemon also creates temporary files and various output files.
If these are filling up the disk, the root folder for temporary files can be changed in Daemon settings.
|

|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
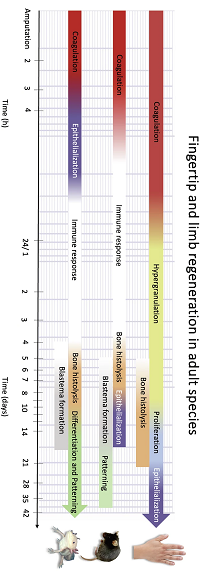
Human fingertip regeneration follows clinical phases with distinct proteomic signatures
Jurek Schultz, Purva A. Patel, Rita Aires, Leah Wissing, Patrick Glatte, Michael Seifert, Marc Gentzel, Guido Fitze, Adele M. Doyle, Tatiana Sandoval-Guzmán
npj Regenerative Medicine volume 10, Article number: 51 (2025), doi:10.1038/s41536-025-00441-y
Human fingertips can regenerate almost fully after distal injuries, but little is known about the regeneration process at a molecular level or how it relates to regenerative animal models.
The authors ran a randomised controlled clinical trial, where a novel silicone finger cap was used for treating fingertip injuries.
Samples of excess wound fluid were collected from the finger cap at regular intervals, covering four distinct clinical phases.
The authors were able to characterise distinct proteomic signatures, processes and active regulatory networks in each phase, as well as show significant divergence from animal models.
The fluid samples were analysed using GelC-MS/MS: separation with SDS-PAGE, tryptic digestion in gel, followed by LC-MS/MS (Thermo Q-Exactive HF) in DDA mode.
After peptide and protein identification with Mascot Server, the results were aggregated in Scaffold.
Low-confidence proteins were filtered out by including only proteins detected in any 3 or more samples, leaving 974 proteins or protein clusters identified in the 36 samples.
Proteins were quantified using spectral counting, followed by PCA, clustering, and pathway and network analysis.
|
|
|
|
 |
|
|
|
Customise the columns in the search log
|
|
|
The Mascot search log is a text file that records useful metadata about database searches (Job#, PID, dbase, User Name, Email, Title, Intermediate file, start time, Duration, Status, Priority, Type, Enzyme, IP address, User ID, Peak list data file).
It can be viewed by following a link from the Mascot front page.
The column display can be configured using little-known configuration options.
The web interface has a one column per checkbox, which controls whether the column is visible (full width) or collapsed (width 2).
To toggle a column, check or uncheck it and click Sort/Filter.
Width and visibility are configurable.
Go to the Mascot configuration editor and select Configuration Options.
Width is controlled by ReviewColWidths 7,8,8,27,30,120,32,25,6,13,4,4,6,16,7,150.
It's measured in characters; for example, the 3rd column is dbase and it defaults to the first 8 characters of the database name.
Visibility is controlled by ReviewColDisplay 1,1,1,1,1,0,0,1,1,1,1,1,1,0,1,1, where 0 means a column is collapsed by default.
In both cases, columns are listed in the fixed order they are shown in the search log.
|

|
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|